Don't wanna be here? Send us removal request.
Text
“Something went wrong”: Disconnect your USB hub to set up a Yubikey in Chrome
I was trying to setup a Yubico FIDO U2F device from a Ubuntu computer, but I couldn't get Google to even get to the pont of allowing me to try. I had set up the .rules file as instructed by the official Yubico site, but that had no effect. Then, I disconnected my USB hub to make sure that there was nothing connected to any USB port. After removing my USB hub, I was able to go through with registration of my Yubikey.
0 notes
Text
Citrix Receiver 13.4 works on Linux
Citrix Receiver after version 13.4 doesn't seem to work on Linux (constant SSL Error 4 messages appear, and Citrix won't load). However, if you revert back to Citrix Receiver 13.4, it will work normally.
0 notes
Text
Bug on iOS 11 allows for the creation of disappearing messages
Obtain an iOS device on iOS 11. Tested on an iPhone with iOS 11.01
Open iMessage and start writing a message
Type the portion of the message that you want to remain visible
Type the "dash" button 3 times. Note: you may not see the cursor advance after each tap
Now type the part of the message you want to have disappear
It seems to be the case that the portion of your message after "---" disappears after your correspondent replies. Enjoy!
(This is probably a bug, but I like it so I'm not reporting it.)
0 notes
Text
Connecting a Teensy 3.2 with an ESP8266
I was having trouble getting a Teensy 3.2 to communicate over Serial1 (pins 0 and 1) with an ESP8266-01S. No baud rate seemed to work: I could see the "ready" message, but there was never a reply to my commands. Recalling that pins 0 and 1 are also touch pins, I moved over to Serial 3 which does not have a touch component. This worked perfectly.
// Forked from https://kaspars.net/blog/electronics/esp8266-teensy-arduino void setup() { // Setup computer to Arduino serial Serial.begin(9600); // Setup Teensy to ESP8266 serial // Use baud rate 115200 during firmware update Serial3.begin(115200); } void loop() { // Send bytes from ESP8266 to computer if ( Serial3.available() ) { Serial.write( Serial3.read() ); } // Send bytes from computer back to ESP8266 if ( Serial.available() ) { Serial3.write( Serial.read() ); delay(100); } }
1 note
·
View note
Text
Installing nvidia-docker on Ubuntu 17.04 (zesty zapus)
docker itself (and nvidia-docker) don't formally support Ubuntu 17.04 Zesty Zapus just yet, but you can still install them successfully like so:
0. Allow apt to use a repository over HTTPS & add Docker's key
sudo apt-get install \ apt-transport-https \ ca-certificates \ curl \ software-properties-common curl -fsSL https://download.docker.com/linux/ubuntu/gpg | sudo apt-key add - # confirm that this matches 9DC8 5822 9FC7 DD38 854A E2D8 8D81 803C 0EBF CD88 sudo apt-key fingerprint 0EBFCD88
1. Install the docker repo for yakkety
sudo add-apt-repository "deb [arch=amd64] https://download.docker.com/linux/ubuntu yakkety stable"
2. Install docker
sudo apt-get update sudo apt-get install docker-ce
3. Install nvidia-docker from a yakkety-compatible source (since there is no zesty-compatible source yet)
cd /tmp wget https://github.com/NVIDIA/nvidia-docker/files/818401/nvidia-docker_1.0.1-yakkety_amd64.deb.zip unzip nvidia-docker_1.0.1-yakkety_amd64.deb.zip sudo dpkg -i /tmp/nvidia-docker*.deb
4. Test run nvidia-docker
nvidia-docker run --rm nvidia/cuda nvidia-smi nvidia-docker run -it -p 8888:8888 gcr.io/tensorflow/tensorflow:latest-gpu
0 notes
Text
SCOTUS of One
With the death of Antonin Scalia, the Supreme Court of the United States (SCOTUS) is down to 8 members. The Senate leaders have gone out of their way to emphasize that they do not plan to give President Obama's nominee, Merrick Garland, a hearing. Rather, they intend to wait until the next president is in office. But what if, in fact, the Senate has realized that it can indefinitely delay the appointment of any future Justice? What if it decides never to give any future nominee a hearing? What if Elena Kagan is the last person ever to be appointed to SCOTUS?
If no future nominees are approved, there will eventually come a day when there is a SCOTUS of One. Who will it be, and when?
I ran 100,000 simulations, based on actuarial data from the National Center for Health Statistics, to determine when each Justice was likely to die based on age, sex, and race. As expected, Elena Kagan lives the longest - her median survival is through year 2046.
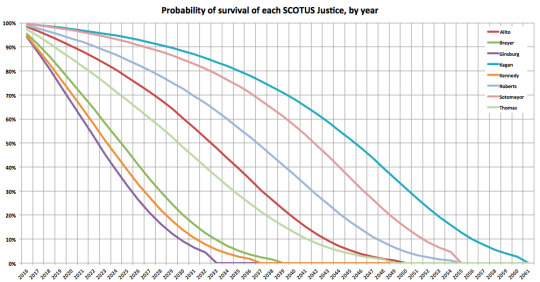
Within each simulation, I kept track of the years where Elena Kagan was the only remaining SCOTUS Justice. This probability peaks in 2047. By that year, Kagan has a 27% chance of being the #SCOTUSofOne.
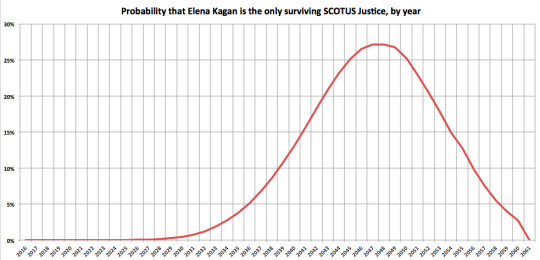
So, the Senate still has time, but the clock is ticking.
(To get higher resolution versions of these images, go to http://imgur.com/a/OEnvc )
0 notes
Text
I installed go with Homebrew on my Mac. Where is the golang standard library?
If you installed golang with homebrew, the path to the go standard library source files is as follows:
/usr/local/Library/LinkedKegs/go/libexec/src/
0 notes
Text
How can I tell if my MacBook SMC reset worked?
Sometimes you need to reset the SMC on your MacBook or MacBook Air - for example, if your batter is no longer charging fully even though the battery itself is working properly. You do this by shutting down the computer, holding down the left shift+ctrl+option buttons and the power button at the same time until the charger flashes from orange to green, then letting go and turning on the computer. It's a bit of a complicated process and it's not obvious when it's done. People often ask "How can I tell if the SMC reset worked?" Here is how you can tell: when you turn you computer back on, open up Activity Monitor, go to the Energy tab, and if "BATTERY (Last 12 hours)" has become a blank slate, the odds are good that the SMC was successfully reset.
0 notes
Quote
People are starting to respond to your new ad set
This is a phrase that you may see when a set of Facebook ads is beginning to get traffic and get clicks. It's a good time to take a look and see if you're getting the response that you're looking for in terms of conversions.
0 notes
Text
Instagram uses an MD5 hash
Unless they are using nonstandard hashes, instagram appears to be using the MD5 hash for their filenames. Take this instagram image, for example:

It comes from https://distilleryimage9.s3.amazonaws.com/aef728f0b82f11e3a0d012d37dc11815_8.jpg "aef728f0b82f11e3a0d012d37dc11815" is a 32-character string of letters from 0-9 and a-f, which is a string representation of a hexadecimal number (base 16 instead of base 10). Thus, this file name requires 128 bits to represent (log(16^32)/log(2)=128). Of the common hash functions, only MD5 yields a 128-bit hash, so MD5 is the most likely answer.
0 notes
Text
Twitter keeps telling me I have notifications, but I don't
This has been happening over the past week: my phone will tell me that I have a new twitter notification. The website will do the same. But when I visit notifications, there's nothing new there.
0 notes
Text
Tell HN: This is why medical regulation exists
The unregulated supplement market is causing irreparable harm.
Half of Americans use supplements at some point, and the supplement market is booming:
Americans spend an estimated $32 billion on dietary supplements every year, attracted by unproven claims that various pills and powders will help them lose weight, build muscle and fight off everything from colds to chronic illnesses. About half of Americans use dietary supplements, and most of them take more than one product at a time.
Thanks in part to a lack of regulation:
But a federal law enacted in 1994, the Dietary Supplement Health and Education Act, prevents the Food and Drug Administration from approving or evaluating most supplements before they are sold.
And it looks like some of these supplements (especially those promoting weight loss) are causing harm:
New data suggests that his is not an isolated case. Dietary supplements account for nearly 20 percent of drug-related liver injuries that turn up in hospitals, up from 7 percent a decade ago
Medical regulation is a balance between safety and progress. With regulation, problems such as these are discovered pre-marketing, while progress seems to be slowed (one might even try to quantitate by how much). Without regulation, problems such as these are discovered post-marketing, when physicians and epidemiologists notice a pattern and correlate it with external trends, (e.g., supplement usage). However, progress (or at least experimentation) occurs more quickly.
It is a trade-off. But this article serves as a reminder that we should be careful before abandoning some of our hard-won regulatory structures wholesale, especially when it comes to the (currently irreplaceable) human body.
0 notes
Text
Sudoku solver in golang
Peter Norvig has a nice article on how to use constraint propagation to solve any soluble Sudoku puzzle, along with code to do so in Python.
I wanted to see how this would look in go, so I copied his technique to create a golang Sudoku solver.
This code takes no advantage of tools such as parallelism (with goroutines), though if the bug strikes again I don't think adding in those features would be particularly challenging.
1 note
·
View note
Text
AskGolang.com: traffic stats, hosting details, and open sourcing the GRAF package
Over the summer before my residency started, I taught my self Go by writing threaded forums with live-loading of comments, called AskGolang. In this post, I give an overview of the traffic from posting that project on Hacker News and #Golang-Nuts; I comment on my hosting; and I talk about how I have now open sourced this project.
Traffic stats
Before open sourcing the project, I posted AskGolang to Hacker News, where it got some interest and spent some time on the front page. I also posted it to the golang-nuts Google group. The site got about 5,000 visitors on that first day:
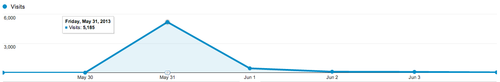
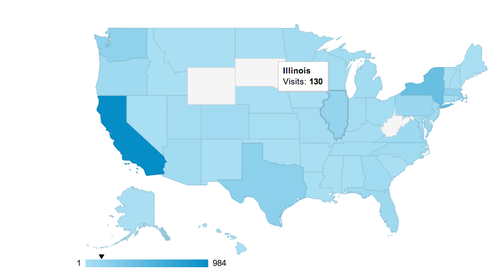
These folks are from all over the US and the world:
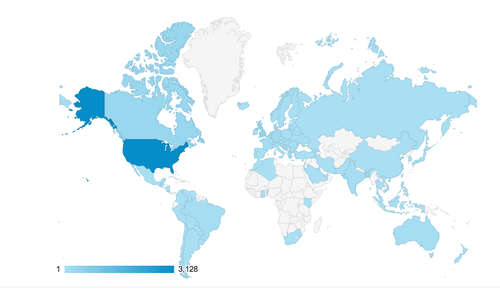
There were a peak of 1,200 visitors within one hour, about one every 3 seconds:
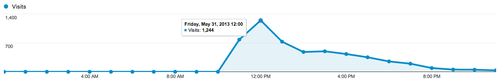
Of these visitors, 134 were sampled by Google Analytics for page speed estimation. On average, the pages loaded in 0.87 second.
As you might imagine, with an average of one visitor per 3 seconds, with pages loading in less than a second, there wasn't much of a chance to run into concurrency roadblocks, and none were noted in the error logs. However, hundreds of users were connected simultaneously with active Websockets. Because I don't have specific logging, I don't have the formal peak number of users, but I was online during this time and the peak was about 120 users simultaneously connected over the same channel via Websockets.
Hosting
People asked about how this project was hosted, and I'm happy to talk about it. My hosting is quite simple: the 1GB (smallest available) Linode running Ubuntu. This costs $20/month, and I host all of my various projects on the same little box. I deploy by asking go to build a Linux binary capable of touching 8 cores, and then I SCP the binary over to the server and tell upstart to reload it. (All of this is automated so I don't skip a step.)
Open source
I've never really participated in open source, except for filing a few bug reports here and there. The code underlying this project is now fully open. While it's still a work very rough and clearly in progress (with console log messages and all!), I wanted to put this out for public consumption now in the hopes of getting feedback that will improve the project and my ability to reason about code. Perhaps, too, someone will use this as a framework to launch a project of their own.
The main "guts" of the project come in the form of a golang package called GRAF, for GRAF Recursively Arranged Forum, a nod to the mechanism for displaying nested comments. That package includes a GRAF example project that is essentially complete (but which requires that you have Postgres installed and that you configure some tables as the forum.sql file dictates).
GRAF, and all of the other components that it uses in various parts, are available as open-source projects available under an MIT-style or an Apache-style license (depending on the project), written by me and/or others.
0 notes
Text
Supreme Court's Myriad decision is deeply confused over cDNA
The Supreme Court just ruled on the Myriad gene patentability case. To recap, Myriad holds patents on the BRCA1 and BRCA2 genes, for which they offer testing to help determine breast cancer risk.
To summarize the decision, the Court held that naturally-occurring DNA, even if it is isolated, is not patentable. However, they also state that "[f]or the reasons that follow," complementary DNA (cDNA) is patentable. What are those reasons?
In section I.A., the court walks through the Central Dogma and does a fine job with it. DNA is transcribed to RNA. RNA gets the introns spliced out and (largely) keeps exons. Processed RNA is then translated by the ribosome into a sequence of amino acids, i.e., a protein. Note that there are a great many functions that RNA serves (tRNA, rRNA, RNA interference, etc). These omissions may become important later. Nevertheless, the Court's description of how genetic information goes from DNA to protein is essentially right. Importantly, they clearly define cDNA as having come from reverse transcription of cDNA. In other words, they correctly define cDNA.
In I.B., the court dismisses Myriad's claims that isolated naturally-occurring BRCA1/2 sequences are patent eligible. Just because you have broken two chemical bonds to free the BRCA1/2 sequence from the rest of the genome, you have not "invented" anything.
Section I.C. starts off with a bang:
cDNA does not present the same obstacles to patentability as naturally occurring, isolated DNA segments. As already explained, creation of a cDNA sequence from mRNA results in an exons-only molecule that is not naturally occurring.
From a scientific standpoint, these opening sentences are hard to reconcile with reality. cDNA naturally occurs quite frequently: it is created by cells infected with retroviruses. The way that we make cDNA is by using reverse transcriptase–which we discovered thanks to retroviruses that exist in the wild–to reverse transcribe RNA to its reverse complement (hence the "c" in cDNA). Is natural cDNA accidentally omitted, intentionally omitted, or is the Court re-defining cDNA to only be artificially reverse transcribed DNA? (Probably not the latter.)
[...] the lab technician unquestionably creates something new when cDNA is made.
Is putting an isolated enzyme (reverse transcriptase) in a test tube with mRNA the "magic juice" that triggers the Court's deference to the technician's "creation"? If it weren't for the splicing work that nature did to remove introns from the mRNA, the technician's work would simply reproduce already-existent DNA, introns and all.
As a result, cDNA is not a “product of nature” and is patent eligible under §101, except insofar as very short series of DNA may have no intervening introns to remove when creating cDNA. In that situation, a short strand of cDNA may be indistinguishable from natural DNA.
Any cDNA may plausibly be found in nature thanks to retroviruses; therefore, literally any cDNA may be indistinguishable from natural DNA.
I am not convinced by the Court's reasoning for permitting cDNA patents on otherwise non-modified genetic sequences. If the Court does not extend this analogy beyond cDNA synthesis, one can imagine ways to show natural occurrence of many important cDNAs, or to alter or create new mRNA sequencing techniques to avoid cDNA intermediates in many RNA applications. However, if the Court extends its analogy beyond the reverse transcription step, then it has stepped into territory that seems quite hazardous for the progress of science and the useful arts.
Follow-up questions
In no particular order:
Would the discovery of naturally-occurring cDNA invalidate a company's patents?
Would the discovery of individuals with gene duplications (that have had introns removed–this happens) invalidate the relevant cDNA patents for that particular gene?
By analogy with the logic here, should any PCR'ed product become patentable, because the technician put an isolated enzyme (DNA polymerase) in a test tube with DNA? (Since the intron splicing step in cDNA construction occurs before the mRNA is reverse transcribed, it's not needed in this analogy.) If not, should any Sanger sequencing product be patentable (since instead of being made entirely of DNA, it's made of DNA plus a modified cap that includes a fluorophor, and therefore differs from the natural DNA template)?
If someone develops a technique that performs artificial DNA intron splicing without a mRNA step, thus yielding DNA indistinguishable from cDNA created in the normal way, should that DNA be patent eligible as if it were cDNA? Should it be considered infringing on patented cDNA of the same sequence?
Should DNA strands created by completely artificial synthesis of DNA–with no RNA step–be patentable? Should they be considered infringing on patented cDNA of the same sequence?
Should we focus on the Court's special emphasis on the reverse transcription of spliced mRNA as the definitive act of human invention? What does this mean for rtPCR? RNA-seq? How can we prevent potential patents on cDNAs in this realm from hindering scientific research?
Will this cause greater interest in mRNA sequencing that avoids a DNA step? One could imagine a variant on Sanger sequencing that uses a mixture of mRNA, non-holoenzyme RNA-dependent RNA polymerases, and RNA nucleotides, along with a small concentration of ddNTPs attached to fluorophores and DNA polymerase for chain termination / sequencing of mRNA without any cDNA stage.
Alternatively, one could imagine a discovered (or artificially created) RNA-dependent RNA polymerase that occasionally incorporates fluorophoric ddNTPs.
Update #1
Recall that the average length of exons in the human genome is >100 nucleotides. Does this ruling mostly harm splice-variant and splice-site research?
2 notes
·
View notes

