#C. F. Møller
Explore tagged Tumblr posts
Text
In the Botta Tradition







Ravnsbjerg Church, Aarhus, Denmark - C. F. Møller
#modern architecture#C. F. Møller#brutalist architecture#masonrydesign#religious architecture#monumental architecture
59 notes
·
View notes
Text

Ravnsbjergkirken (1976) in Aarhus, Denmark, by C. F. Møllers Tegnestue
1K notes
·
View notes
Text

Ravnsbjerg Church. Aarhus, Denmark
Architect: C. F. Møller, 1976
1 note
·
View note
Text
School of Business and Social Sciences, Aarhus University
Aarhus University BSS, Danish Architecture Competition, Jutland Building Project, Images
School of Business and Social Sciences in Aarhus, Jutland
30 Apr 2021
School of Business and Social Sciences
Design: AART
Location: Århus, Jylland, Danmark
The Danish practice AART has won the competition to design the new Aarhus BSS, School of Business and Social Sciences at Aarhus University, which is the second largest university in Denmark. Flexible study environments and sustainable solutions combined with high architectural qualities. These are the key elements in the future Aarhus BSS.
The ambition is to create a new landmark for the city of Aarhus and a meeting point in the University City – the upcoming city-integrated campus at Aarhus University. Particularly significant will be the large auditorium, which will be established as a rotunda in glass and aluminium in the central square and thus become a spectacular centre of the campus.
In 2025, Aarhus BSS will move its activities across the city – from its current address to the University City. With the move, students and staff can, among other things, can look forward to a number of new and larger auditoriums – where one of them will be Aarhus University’s largest with room for 800 people. Wood will be a common material in the architectural expressions of the new buildings, while study areas and teaching rooms are designed so that they can be used for several purposes.
This applies, for example, to the large auditorium, which can be divided into both two and three smaller auditoriums and used for both lectures and external events. A flexibility that is also worked into the teaching rooms, which in all buildings become a well-integrated part of the study environments.
“The project is ambitious and supports the overall plan’s visions for the development of the area as a sustainable urban area that will be full of life 24/7”, says Jørgen Lang, CEO of FEAS.
With its location in the middle of the University City and its 37,000 square meters, Aarhus BSS will turn into an important part of Aarhus University’s overall physical development where all new buildings will be closer to the faculty’s other activities in the University Park in Aarhus. With its architectural expression and quality, it manages to create and clear coherence across the University Park and the University City. That is why it is moving towards another simply unified and integrated campus in Aarhus, where the synergies strengthen between research environments, study environments, teaching, and collaboration with the university’s external stakeholders.
Last but not least, the sustainable ambitions are high for the future Aarhus BSS. Both active and passive sustainable solutions have therefore been entered from the start of the project, which aims for a DGNB Gold certification – on a par with the specification that the University City as an urban area has achieved.
Aarhus BSS in the University City – Building Information
Architects: AART Location: Aarhus, Denmark Size: 37,000 square meters Developer: FEAS Landscape architect: LYTT Architecture Entrepreneur: A. Enggaard
School of Business and Social Sciences, Aarhus University images / information received 300421
Location: Aarhus, Jutland, Denmark
Architecture in Denmark
Jutland Architectural Projects
Aarhus Architecture Designs – chronological list
Aarhus Architecture Walking Tours
Home for Life, Lystrup Design: AART Architects photograph : Adam Mørk Home for Life, Denmark
Pop-up Train Carriages in Aarhus Design: AART Architects image from architecture studio Pop-up Train Carriages in Aarhus
Nicolinehus Aarhus Ø Design: AART Architects image from architect Nicolinehus Aarhus Ø
Danish Architecture
Danish Architect
Denmark Architecture
Aarhus Concert Hall by Arkitektfirmaet C. F. Møller:
Utzon Center, Aalborg, Jylland Kim Utzon Architects photo : Torben Eskerod Denmark
Århus harbour – docklands building UNStudio & 3XN Architects Aarhus Harbour, 2022, UN Studio
The Iceberg Project Aarhus : Julien De Smedt Architects
Contemporary Housing
Aarhus Building : Further Information re Harbour project
Contemporary Housing Designs
C.F. Møller Architects
Comments / photos for the School of Business and Social Sciences, Aarhus University page welcome
Website: Aarhus
The post School of Business and Social Sciences, Aarhus University appeared first on e-architect.
1 note
·
View note
Photo

Kay Fisker, Svenn Eske Kristensen og C. F. Møller. Dronningegården, Copenhagen, 1943.
62 notes
·
View notes
Note
mesaj attım ama cevap vermedin dostum. PCR ve qPCR teknikleriyle alakalı kaynak olarak yardımcı olabilir misin?
mendeley kullanıyor musun bilmiyorum ama ben APA şeklinde buraya atayım kaynakları sen seç beğen al dostum.
Polymerase chain reaction - Wikipedia. (n.d.). Retrieved May 27, 2020, from https://en.wikipedia.org/wiki/Polymerase_chain_reaction
Citations for Chemical Breakthrough Awards 2017 Awardees. (n.d.). Division of the History of Chemistry. Retrieved May 27, 2020, from http://www.scs.illinois.edu/~mainzv/HIST/awards/CCB-2017_Awardees.php
Kary B. Mullis – Nobel Lecture: The Polymerase Chain Reaction. (n.d.). Retrieved May 27, 2020, from http://nobelprize.org/nobel_prizes/chemistry/laureates/1993/mullis-lecture.html
Advice on How to Survive the Taq Wars. (2006). GEN Genetic Engineering News – Biobusiness Channel, 26(9). https://www.genengnews.com/magazine/49/advice-on-how-to-survive-the-taq-wars/
Babineaux, M. J., & Alter, M. J. (2011). Viral Hepatitis. In Netter’s Infectious Disease (pp. 399–410). Elsevier Inc. https://doi.org/10.1016/B978-1-4377-0126-5.00067-7
Key ingredient in coronavirus tests comes from Yellowstone’s lakes. (2020). Science. https://www.nationalgeographic.com/science/2020/03/key-ingredient-in-coronavirus-tests-comes-from-yellowstone/
Bing, D., Boles, C., Rehman, F., Audeh, M., Belmarsh, M., Kelley, B., & Adams, C. (1996). Bridge amplification: a solid phase PCR system for the amplification and detection of allelic differences in single copy genes. Genetic Identity Conference Proceedings, Seventh International Symposium on Human Identification. http://www.promega.com/geneticidproc/ussymp7proc/0726.html
Tomar, R. S. (2010). Molecular markers & plant biotechnology. New India Pub. Agency.
Full Text – LaNe RAGE: a new tool for genomic DNA flanking sequence determination. (n.d.). Retrieved May 27, 2020, from http://www.ejbiotechnology.info/content/vol8/issue2/full/7/index.html
Kieleczawa, J. (2006). DNA sequencing II : optimizing preparation and cleanup. Jones and Bartlett Publishers.
David, F. (2002). Utiliser les propriétés topologiques de l’ADN: une nouvelle arme contre les agents pathogènes. Fusion. http://www.lab-rech-associatives.com/pdf/Utiliser la Topologie de l’ADN.pdf
Kellogg, D. E., Rybalkin, I., Chen, S., Mukhamedova, N., Vlasik, T., Siebert, P. D., & Chenchik, A. (1994). TaqStart antibody(TM): “Hot start” PCR facilitated by a neutralizing monoclonal antibody directed against Taq DNA polymerase. BioTechniques, 16(6), 1134–1137.
Ochman, H., Gerber, A. S., & Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics, 120(3), 621–623.
Ninfa, A. B. et all. (2010). Fundamental laboratory Approaches for Biochemistry and Biotechnology about PAGE. In Fundamental laboratory Approaches for Biochemistry and Biotechnology. John Wiley & Sons. inc.
Boehnke, M., Arnheim, N., Li, H., & Collins, F. S. (1989). Fine-structure genetic mapping of human chromosomes using the polymerase chain reaction on single sperm: Experimental design considerations. American Journal of Human Genetics, 45(1), 21–32.
Coronavirus: il viaggio dei test. (n.d.). Istituto Superiore Di Sanità. Retrieved May 27, 2020, from https://www.iss.it/web/guest/primo-piano/-/asset_publisher/o4oGR9qmvUz9/content/id/5269706
Patidar, M., Agrawal, S., Parveen, F., & Khare, P. (2015). Molecular insights of saliva in solving paternity dispute. Journal of Forensic Dental Sciences, 7(1), 76. https://doi.org/10.4103/0975-1475.150325
Chemical Synthesis, Sequencing, and Amplification of DNA (class notes on MBB/BIO 343). (n.d.). Arizona State University. Retrieved May 27, 2020, from http://photoscience.la.asu.edu/photosyn/courses/BIO_343/lecture/DNAtech.html
Electronic PCR. (n.d.). NCBI – National Center for Biotechnology Information. Retrieved May 27, 2020, from https://www.ncbi.nlm.nih.gov/sutils/e-pcr/
Bogetto, P. and W. L. (2000). Helpful tips for PCR. Focus, 22(1), 12. http://tools.thermofisher.com/Content/Focus/Focus Volume 22 Issue 1.pdf
Mullis, K. B. (1990). The unusual origin of the polymerase chain reaction. Scientific American, 262(4), 56–65. https://doi.org/10.1038/scientificamerican0490-56
Borman, J., Schuster, D., Li, W., Jessee, J., & Rashtchian, A. (2000). PCR from problematic templates. Focus, 22(1), 10. http://tools.thermofisher.com/Content/Focus/Focus Volume 22 Issue 1.pdf
Mullis, K. B. (1998). Dancing naked in the mind field. Pantheon Books. https://archive.org/details/dancingnakedinmi00mull
PCR. (n.d.). Genetic Science Learning Center, University of Utah. Retrieved May 27, 2020, from http://learn.genetics.utah.edu/content/labs/pcr/
Polymerase Chain Reaction (PCR). (n.d.). National Center for Biotechnology Information, U.S. National Library of Medicine. Retrieved May 27, 2020, from https://www.ncbi.nlm.nih.gov/probe/docs/techpcr/
Rabinow, P. (1996). Making PCR : a story of biotechnology. University of Chicago Press. https://archive.org/details/makingpcrstoryof00rabi
Park, D. J. (2005). A new 5′ terminal murine GAPDH exon identified using 5′RACE LaNe. Molecular Biotechnology, 29(1), 39–46. https://doi.org/10.1385/MB:29:1:39
White, B. A. (1993). PCR protocols : current methods and applications. Humana Press.
Myrick, K. V., & Gelbart, W. M. (2002). Universal Fast Walking for direct and versatile determination of flanking sequence. Gene, 284(1–2), 125–131. https://doi.org/10.1016/S0378-1119(02)00384-0
McPherson, M. J., & Møller, S. G. (2006). PCR. Taylor & Francis.
Bagchi, D. (2013). Bio-nanotechnology : a revolution in food, biomedical, and health sciences. Wiley-Blackwell.
Liu, Y. G., & Whittier, R. F. (1995). Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics, 25(3), 674–681. https://doi.org/10.1016/0888-7543(95)80010-J
Thornhill, A. (2007). Single Cell Diagnostics. Cell, 132. https://doi.org/10.1007/978-1-59745-298-4
Khan, Z., Poetter, K., & Park, D. J. (2008). Enhanced solid phase PCR: mechanisms to increase priming by solid support primers. Analytical Biochemistry, 375(2), 391–393. https://doi.org/10.1016/j.ab.2008.01.021
Baron, S. (1996). Medical microbiology (4th ed.). University of Texas Medical Branch at Galveston. https://www.ncbi.nlm.nih.gov/books/NBK7813/
Bustin, S. A., & Nolan, T. (2009). Analysis of mRNA Expression by Real-time. In Real-Time PCR: Current Technology and Applications. Caister Academic Press. http://www.caister.com/realtimepcr
White, B. A., Shyamala, V., & Ames, G. F.-L. (2003). Single Specific Primer-Polymerase Chain Reaction (SSP-PCR) and Genome Walking. In PCR Protocols (pp. 339–348). Humana Press. https://doi.org/10.1385/0-89603-244-2:339
Dobosy, J. R., Rose, S. D., Beltz, K. R., Rupp, S. M., Powers, K. M., Behlke, M. A., & Walder, J. A. (2011). RNase H-dependent PCR (rhPCR): Improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnology, 11, 80. https://doi.org/10.1186/1472-6750-11-80
Sambrook, J., & Russell, D. W. (David W. (2001). Molecular cloning : a laboratory manual (3rd ed.). Cold Spring Harbor Laboratory Press. https://archive.org/details/molecularcloning0000samb_p7p5
David, F., & Turlotte, É. (1998). Une methode d’amplification genique isotherme. Comptes Rendus de l’Academie Des Sciences - Serie III, 321(11), 909–914. https://doi.org/10.1016/S0764-4469(99)80005-5
Shen, C., & Zhang, Z. (2013). An Overview of Nanoparticle-Assisted Polymerase Chain Reaction Technology. In Bio-Nanotechnology: A Revolution in Food, Biomedical and Health Sciences (pp. 97–106). Blackwell Publishing Ltd. https://doi.org/10.1002/9781118451915.ch5
Kleppe, K., Ohtsuka, E., Kleppe, R., Molineux, I., & Khorana, H. G. (1971). Studies on polynucleotides. XCVI. Repair replication of short synthetic DNA’s as catalyzed by DNA polymerases. Journal of Molecular Biology, 56(2), 341–361. https://doi.org/10.1016/0022-2836(71)90469-4
Park, D. J. (2004). 3′ RACE LaNe: A simple and rapid fully nested PCR method to determine 3′-terminal cDNA sequence. BioTechniques, 36(4), 586–590. https://doi.org/10.2144/04364bm04
Kim, B., Kim, J., Kim, M., Kim, B.-D., Jo, S., Park, S., Son, J., Hwang, S., Dugasani, S., Chang, I., Kim, M., & Liu, W. (2016). Ternary and senary representations using DNA double-crossover tiles. Nanotechnology, 10(45), 105601. https://doi.org/10.1088/0957-4484
Isenbarger, T. A., Finney, M., Ríos-Velázquez, C., Handelsman, J., & Ruvkun, G. (2008). Miniprimer PCR, a new lens for viewing the microbial world. Applied and Environmental Microbiology, 74(3), 840–849. https://doi.org/10.1128/AEM.01933-07
Raoult, D., Aboudharam, G., Crubézy, E., Larrouy, G., Ludes, B., & Drancourt, M. (2000). Molecular identification by “suicide PCR” of Yersinia pestis as the agent of Medieval Black Death. Proceedings of the National Academy of Sciences of the United States of America, 97(23), 12800–12803. https://doi.org/10.1073/pnas.220225197
Herman, J. G., Graff, J. R., Myöhänen, S., Nelkin, B. D., & Baylin, S. B. (1996). Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America, 93(18), 9821–9826. https://doi.org/10.1073/pnas.93.18.9821
Zietkiewicz, E., Rafalski, A., & Labuda, D. (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics, 20(2), 176–183. https://doi.org/10.1006/geno.1994.1151
San Millán, R. M., Martínez-Ballesteros, I., Rementeria, A., Garaizar, J., & Bikandi, J. (2013). Online exercise for the design and simulation of PCR and PCR-RFLP experiments. BMC Research Notes, 6(1), 513. https://doi.org/10.1186/1756-0500-6-513
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., & Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene, 77(1), 61–68. https://doi.org/10.1016/0378-1119(89)90359-4
Schwartz, J. J., Lee, C., & Shendure, J. (2012). Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nature Methods, 9(9), 913–915. https://doi.org/10.1038/nmeth.2137
Mueller, P. R., & Wold, B. (1989). In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science, 246(4931), 780–786. https://doi.org/10.1126/science.2814500
Priye, A., Hassan, Y. A., & Ugaz, V. M. (2013). Microscale chaotic advection enables robust convective DNA replication. Analytical Chemistry, 85(21), 10536–10541. https://doi.org/10.1021/ac402611s
Vincent, M., Xu, Y., & Kong, H. (2004). Helicase-dependent isothermal DNA amplification. EMBO Reports, 5(8), 795–800. https://doi.org/10.1038/sj.embor.7400200
Pierce, K. E., & Wangh, L. J. (2007). Linear-After-The-Exponential Polymerase Chain Reaction and Allied Technologies (pp. 65–85). https://doi.org/10.1007/978-1-59745-298-4_7
Krishnan, M., Ugaz, V. M., & Burns, M. A. (2002). PCR in a Rayleigh-Bénard convection cell. Science, 298(5594), 793. https://doi.org/10.1126/science.298.5594.793
Innis, M. A., Myambo, K. B., Gelfand, D. H., & Brow, M. A. D. (1988). DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proceedings of the National Academy of Sciences of the United States of America, 85(24), 9436–9440. https://doi.org/10.1073/pnas.85.24.9436
Stemmer, W. P. C., Crameri, A., Ha, K. D., Brennan, T. M., & Heyneker, H. L. (1995). Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene, 164(1), 49–53. https://doi.org/10.1016/0378-1119(95)00511-4
Alonso, A., Martín, P., Albarrán, C., García, P., García, O., Fernández De Simón, L., García-Hirschfeld, J., Sancho, M., De La Rúa, C., & Fernández-Piqueras, J. (2004). Real-time PCR designs to estimate nuclear and mitochondrial DNA copy number in forensic and ancient DNA studies. Forensic Science International, 139(2–3), 141–149. https://doi.org/10.1016/j.forsciint.2003.10.008
Yeh, S. H., & Mink, C. A. M. (2012). Bordetella pertussis and Pertussis (Whooping Cough). In Netter’s Infectious Disease (pp. 11–14). Elsevier Inc. https://doi.org/10.1016/B978-1-4377-0126-5.00003-3
Kwok, S., Mack, D. H., Mullis, K. B., Poiesz, B., Ehrlich, G., Blair, D., Friedman-Kien, A., & Sninsky, J. J. (1987). Identification of human immunodeficiency virus sequences by using in vitro enzymatic amplification and oligomer cleavage detection. Journal of Virology, 61(5), 1690–1694. https://doi.org/10.1128/jvi.61.5.1690-1694.1987
Cai, H. Y., Caswell, J. L., & Prescott, J. F. (2014). Nonculture Molecular Techniques for Diagnosis of Bacterial Disease in Animals: A Diagnostic Laboratory Perspective. In Veterinary Pathology (Vol. 51, Issue 2, pp. 341–350). https://doi.org/10.1177/0300985813511132
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J., & Wittwer, C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622. https://doi.org/10.1373/clinchem.2008.112797
Quill, E. (2008). Medicine: Bood-matching goes genetic. In Science (Vol. 319, Issue 5869, pp. 1478–1479). https://doi.org/10.1126/science.319.5869.1478
Garibyan, L., & Avashia, N. (2013). Polymerase chain reaction. Journal of Investigative Dermatology, 133(3), 1–4. https://doi.org/10.1038/jid.2013.1
Movert, E., Wu, Y., Lambeau, G., Kahn, F., Touqui, L., & Areschoug, T. (2013). Using Patient Pathways to Accelerate the Drive to Ending Tuberculosis. Journal of Infectious Diseases, 208(12), 2025–2035. https://doi.org/10.1093/INFDIS
Lawyer, F. C., Stoffel, S., Saiki, R. K., Chang, S. Y., Landre, P. A., Abramson, R. D., & Gelfand, D. H. (1993). High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5′ to 3′ exonuclease activity. Genome Research, 2(4), 275–287. https://doi.org/10.1101/gr.2.4.275
Pombert, J. F., Sistek, V., Boissinot, M., & Frenette, M. (2009). Evolutionary relationships among salivarius streptococci as inferred from multilocus phylogenies based on 16S rRNA-encoding, recA, secA, and secY gene sequences. BMC Microbiology, 9, 232. https://doi.org/10.1186/1471-2180-9-232
Chien, A., Edgar, D. B., & Trela, J. M. (1976). Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. Journal of Bacteriology, 127(3), 1550–1557. https://doi.org/10.1128/jb.127.3.1550-1557.1976
Sharkey, D. J., Scalice, E. R., Christy, K. G., Atwood, S. M., & Daiss, J. L. (1994). Antibodies as thermolabile switches: High temperature triggering for the polymerase chain reaction. Bio/Technology, 12(5), 506–509. https://doi.org/10.1038/nbt0594-506
Carr, A. C., & Moore, S. D. (2012). Robust quantification of polymerase chain reactions using global fitting. PLoS ONE, 7(5), e37640. https://doi.org/10.1371/journal.pone.0037640
Rhizobium, G. E. (2013). Complete Genome Sequence of the Sesbania Symbiont and Rice. Nucleic Acids Research, 1(1256879), 13–14. https://doi.org/10.1093/nar
Pavlov, A. R., Pavlova, N. V., Kozyavkin, S. A., & Slesarev, A. I. (2004). Recent developments in the optimization of thermostable DNA polymerases for efficient applications. In Trends in Biotechnology (Vol. 22, Issue 5, pp. 253–260). https://doi.org/10.1016/j.tibtech.2004.02.011
Determining Annealing Temperatures for Polymerase Chain Reaction. (2012). The American Biology Teacher, 74(4), 256–260. https://doi.org/10.1525/abt.2012.74.4.9
Cheng, S., Fockler, C., Barnes, W. M., & Higuchi, R. (1994). Effective amplification of long targets from cloned inserts and human genomic DNA. Proceedings of the National Academy of Sciences of the United States of America, 91(12), 5695–5699. https://doi.org/10.1073/pnas.91.12.5695
Saiki, R., Gelfand, D., Stoffel, S., Scharf, S., Higuchi, R., Horn, G., Mullis, K., & Erlich, H. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239(4839), 487–491. https://doi.org/10.1126/science.239.4839.487
Saiki, R. K., Scharf, S., Faloona, F., Mullis, K. B., Horn, G. T., Erlich, H. A., & Arnheim, N. (1985). Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 230(4732), 1350–1354. https://doi.org/10.1126/science.2999980
2 notes
·
View notes
Photo

C. F. Møllerがコンペに勝利し、デンマーク、VIA University Collegeの新キャンパスを設計することに (ArchDaily) C. F. Møller’s Competition Winning design for VIA University College in Denmark (ArchDaily)
0 notes
Photo

StadtBauwelt issue 19.2017 “Is the High-Rise the Future of Housing?” published on September 22nd, 2017
#bauwelt magazine#german architectural magazine#architectural magazine#bauwelt#stadtbauwelt#issue 19.2017#editorial office bauwelt#editorial office berlin#high-rise#housing#malte saenger#guenter vornholz#hamonic + masson & associés#comte vollenweider#LIN Architekten und Urbanisten#c. f. møller#LAN#lacaton vassal#Meili Peter Architekten#pysall architekten#buchner bründler#cobe#alexanderplatz
0 notes
Photo


Vestersøhus housing block Copenhagen, Denmark; 1935-39
Kay Fisker, C. F. Møller
see map | more information 1, 2, 3
via "Das Werk" 35 (1948)
#architecture#arquitectura#architektur#architettura#vestersohus#vestersøhus#apartment#apartment building#kay fisker#cf moller#copenhagen#denmark#danish architecture
56 notes
·
View notes
Text

Canada Soccer’s Women’s National Team will launch their all-important 2019 season with two matches in Spain including a training match against Switzerland on 17 January and an international friendly match against Norway on 22 January in La Manga.
Canada will also compete in its eighth Algarve Cup which kicks off in February. Drawn into Group A, Canada will face Iceland on 27 February and Scotland on 1 March before playing a final match on 6 March to determine its final rank in the tournament. Canada won the Algarve Cup in 2016 and placed second in 2017.
Seven of the 12 teams competing in the 2019 Algarve Cup are headed to the FIFA Women’s World Cup France 2019™ including China, Norway, Scotland, Spain, Sweden, and Group E opponents Canada and the Netherlands. The other five participating teams are Demark, Iceland, Poland, hosts Portugal, and Switzerland.
Canada’s first two matches of 2019 mark the start of its FIFA Women’s World Cup France 2019™ campaign which kicks off Monday 10 June when they face Cameroon at Stade de la Mosson in Montpellier, France. Canada will then travel to Grenoble for their second group match on Saturday 15 June against New Zealand at Stade des alpes. Canada then conclude the group phase in Reims on Thursday 20 June with a match against Netherlands at Stade August-Velaune.
“This year is all about preparing for the FIFA Women’s World Cup, so we are eager to get together in Europe and continue the momentum we built throughout 2018,” said Kenneth Heiner-Møller, Canada Soccer’s Women’s National Team Head Coach. “Switzerland and Norway are both difficult sides, so these two matches will serve as an excellent opportunity to test ourselves against strong European opponents.”
While Switzerland missed out on the 2019 edition of the FIFA Women’s World Cup, a strong performance brought them to the Round of 16 at the FIFA Women’s World Cup Canada 2015™ where they fell to hosts Canada 1:0. Norway will be preparing for its own FIFA Women’s World Cup campaign, having been drawn into Group A against hosts France, Korea Republic and Nigeria. Norway qualified for the FIFA Women’s World Cup France 2019™ after defeating the Netherlands 2-1, who Canada will face in the 2019 group stage, in their final group game of UEFA World Cup Qualifying.
“Heading into our first camp of 2019 we are working to establish a strong foundation from which we can build on over the next few months heading into the FIFA Women’s World Cup,” said Heiner-Møller. “We are certainly not starting from scratch, this team has been developed over time, but work remains to ensure that when we take the pitch in June, Canada will be at their very best, putting on a genuine FIFA World Cup performance.”
January 2019 Roster
Canada’s January 2019 match roster combines team veterans with standout young players. Captain Christine Sinclair will lead the team as she continues to close in on the all-time FIFA scoring record needing just seven goals to earn the top spot. Teenagers including Jordyn Huitema and Jayde Riviere, both 17 years old, recently helped Canada earn a fourth-place finish at the FIFA U-17 Women’s World Cup Uruguay 2018 and will be joined by Julia Grosso (age 18) and Deanne Rose (age 19).
Ottawa’s Vanessa Gilles will be making her first appearance in a Canada Soccer National Team camp.
Canada Soccer Women’s National Team January 2019 Roster
#1 GK Stephanie Labbe, age 32, from Stony Plain, AB/ Linköpings FC (Damallsvenskan)
#2 FB Allysha Chapman, age 29, from Courtice, ON/ Houston Dash (NWSL)
#3 CB Kadeisha Buchanan, age 23, from Brampton, ON/ Olympique Lyonnais (Division 1 Féminine France)
#4 CB Shelina Zadorsky, age 26, from London, ON/ Orlando Pride (NWSL)
#5 M Rebecca Quinn, age 23, from Toronto, ON/ Washington Spirit (NWSL)
#6 F Deanne Rose, age 19, from Alliston, ON/ University of Florida Gators (NCAA)
#7 M Julia Grosso, age 18, from Vancouver, BC / Vancouver Whitecaps FC Girls Elite/ Canada Soccer Regional EXCEL Super Centre (British Columbia)
#9 F Jordyn Huitema, age 17, from Chilliwack, BC/ Vancouver Whitecaps FC Girls Elite/ Canada Soccer Regional EXCEL Super Centre (British Columbia)
#10 FB Ashley Lawrence, age 23, from Caledon, ON/Paris Saint Germain (Division 1 Féminine France)
#11 M Desiree Scott, age 31, from Winnipeg, MB/ Utah Royals FC (NWSL)
#12 F Christine Sinclair ( C ), age 35, from Burnaby, BC/ Portland Thorns (NWSL)
#13 M Sophie Schmidt, age 30, from Abbotsford, BC
#15 F Nichelle Prince, age 23, from Ajax, ON/ Houston Dash (NWSL)
#16 F Janine Beckie, age 24, from Highlands Ranch, CO/ Manchester City (FA Women's Super League)
#17 M Jessie Fleming, age 20, from London, ON/UCLA (NCAA)
#18 GK Sabrina D'Angelo, age 25, from Welland, ON/North Carolina Courage (NWSL)
#19 F Adriana Leon, age 26, from King City, ON
#20 FB Shannon Woeller, age 28, from Vancouver, BC/ Eskilstuna United DFF (Damallsvenskan)
#21 GK Kailen Sheridan, age 23, from Whitby, ON/ Sky Blue FC (NWSL)
#22 FB Lindsay Agnew, age 23, from Kingston, ON/ Houston Dash (NWSL)
#23 FB Jayde Riviere, age 17, from Markham, ON/ Vancouver Whitecaps FC Girls Elite/ Canada Soccer Regional EXCEL Super Centre (Ontario)
#24 FB/F Jenna Hellstrom, age 23, from Sudbury, ON/ Växjö DFF (Damallsvenskan)
#25 CB Vanessa Gilles, 22, from Ottawa, ON/ Les Girondins de Bordeaux (Division 1 Féminine France)
Canada Soccer Women’s National Team 2019 Match Schedule
*Additional matches to be announced in Spring 2019.
Thursday 17 January – Canada vs Switzerland (Closed door training match)
Tuesday 22 January – Canada vs Norway – 18:00 local (12:00 ET/09:00 PT) at La Manga Stadium in La Manga, Spain (Closed door, international friendly match)
2019 Algarve Cup
Wednesday 27 February – Canada vs Iceland (Time and location TBC)
Friday 1 March – Canada v Scotland (Time and location TBC)
Wednesday 6 March – Canada v TBD (Time and location TBC)
23 notes
·
View notes
Text
To mænd forsøgte fredag eftermiddag kl. 14.08 at flygte fra politiet i en bil på C. F. Møllers Alle i Aarhus - det var en rigtig dårlig idé. Civilklædt politi havde kort forinden observeret, at der foregik salg af narkotika fra en bilen. De rettede henvendelse til føreren af bilen, som i stedet satte i gang med hjulspind og kørte frem mod en politibetjent som måtte springe til side for ikke at blive påkørt. Yderligere måtte flere personer springe til side for ikke at blive påkørt af den flygtende narkopusher. Kort efter standsede bilen dog op og en passagerer løb fra bilen. Føreren, en 24 årig mand, blev anholdt i sin bil og indbragt til Politigården. Den 24 årige mand sigtet for at have forvoldt fare for liv og førlighed, salg af narkotika, narkokørsel, flere færdselslovsovertrædelser. Politiet har ligeledes beslaglagt bilen, da hans forseelser betragtes som vanvidskørsel. Den 24-årige er løsladt efter endt afhøring - og bilen er bragt til politiets lager af beslaglagte biler.
0 notes
Text
F-box proteins typically recruit target proteins to the SCF complex so that they can be tagged with multiple copies of ubiquitin by the E3 ligase (see Figure 15.33A). (...) Several of these F-box proteins function as hormone receptor complexes (Figure 15.33B and C). (...) In the presence of auxin, AUX/IAA repressors are recruited to the TIR1/AFB receptor complex and are tagged with ubiquitin for degradation by the 26S proteasome (see Figure 15.33B).
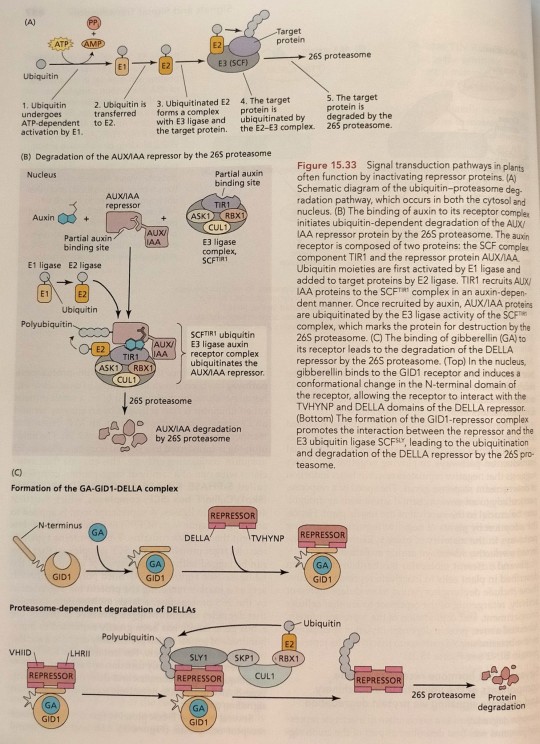
"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quotes#plant physiology and development#nonfiction#textbook#f box#scf complex#ubiquitin#ligase#hormones#plant hormones#hormone receptors#auxin#aux#iaa#auxin indole acetic acid#afb#tir1#proteasome
0 notes
Text

Møllevangskirken (1958-59) in Aarhus, Denmark, by C. F. Møller
158 notes
·
View notes
Text

Ravnsbjerg Church. Aarhus, Denmark
Architect: C. F. Møller, 1976
0 notes
Text
The plant hormones jasmonate and gibberellin also promote the interaction between an F-box protein of a SCF ubiquitin E3 ligase and its transcriptional repressor target proteins (Figure 15.34B and C).
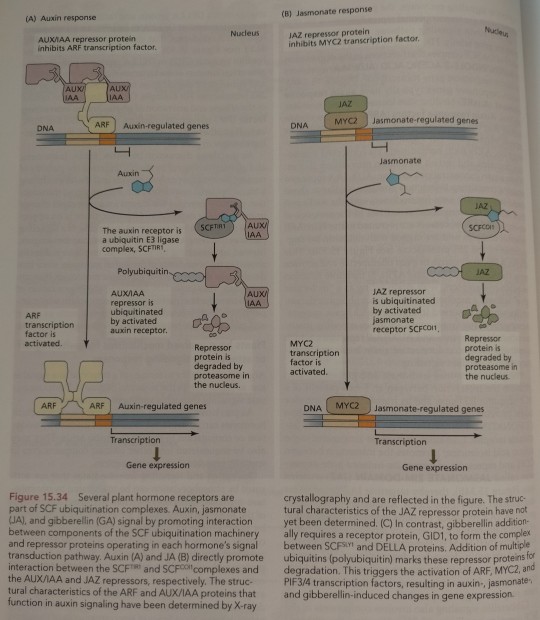
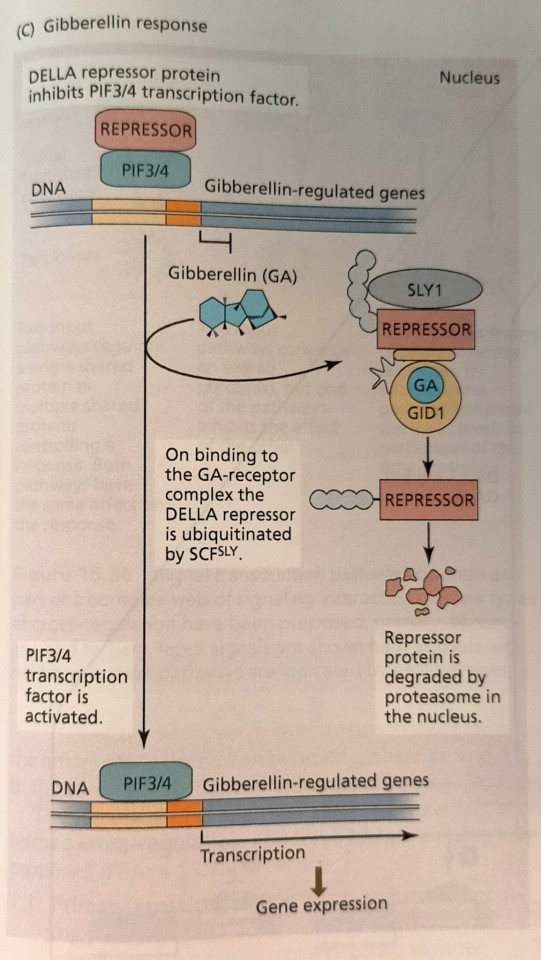
"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quote#plant physiology and development#nonfiction#textbook#hormones#plant hormones#jasmonate#gibberellin#f box protein#ubiquitin#ligase
0 notes
Photo

Aarhus University
Architects. Kay Fisker, C. F. Møller and Povl Stegmann
Aarhus, Denmark
January, 2018
Tumblr - Instagram
184 notes
·
View notes