#techniques in molecular systematics and evolution
Explore tagged Tumblr posts
Text
How Biological Research is Changing the World
Introduction
Biological research is the backbone of scientific advancements, driving innovations in medicine, genetics, biotechnology, and environmental science. It involves the systematic study of living organisms, their functions, and their interactions to understand life at the molecular and cellular levels. As researchers continue to explore the vast complexities of biology, new discoveries are revolutionizing healthcare, agriculture, and sustainability.
Understanding Biological Research
Biological research is a multidisciplinary field that investigates the structure, function, growth, and evolution of living organisms. Scientists use various experimental methods and technologies to uncover the underlying mechanisms that govern life. The insights gained from biological research contribute to medical breakthroughs, environmental conservation, and biotechnological advancements.
Importance of Biological Research in Modern Science
Biological research plays a crucial role in addressing global challenges, from combating diseases to improving agricultural productivity. It has led to the development of life-saving vaccines, advanced genetic therapies, and innovative farming techniques. Additionally, biological research aids in understanding climate change, biodiversity conservation, and ecological balance, ensuring a sustainable future for generations to come.
Key Fields of Biological Research
Biological research encompasses various disciplines, each contributing to scientific progress in unique ways:
Genetics and Genomics – This field focuses on understanding DNA, hereditary diseases, and genetic engineering to develop targeted therapies.
Microbiology and Immunology – Research in these areas helps in studying microorganisms, disease-causing pathogens, and the human immune response to infections.
Biotechnology and Bioengineering – This involves applying biological principles to develop medical treatments, industrial solutions, and agricultural improvements.
Environmental Biology – Researchers in this field study ecosystems, climate change, and ways to protect biodiversity.
Cutting-Edge Techniques in Biological Research
Technological advancements have significantly improved biological research methodologies. Some of the most impactful techniques include:
CRISPR Gene Editing – A revolutionary tool that allows scientists to modify DNA sequences with precision, potentially curing genetic disorders.
Next-Generation Sequencing (NGS) – A high-throughput technology used to decode genetic material quickly, aiding in cancer research and personalized medicine.
Artificial Intelligence in Biology – AI-powered data analysis enhances research accuracy, speeds up drug discovery, and predicts disease patterns.
Role of Biological Research in Medicine and Healthcare
Biological research has transformed healthcare by introducing innovative treatments and diagnostic tools. It has led to the development of vaccines, such as those for COVID-19, and has played a vital role in cancer immunotherapy. Personalized medicine, which tailors treatments based on an individual's genetic profile, is also a direct outcome of extensive biological research.
Ethical Considerations in Biological Research
As scientific advancements progress, ethical concerns arise, particularly in genetic modifications, cloning, and biodiversity conservation. The responsible use of genetic engineering technologies, such as CRISPR, requires careful regulation to prevent unintended consequences. Ethical research practices ensure that advancements benefit humanity without causing harm to ecosystems or individuals.
Future Prospects of Biological Research
The future of biological research holds immense potential, with advancements in biotechnology, regenerative medicine, and AI-driven research leading the way. Scientists continue to explore ways to enhance human health, improve food security, and develop sustainable solutions for environmental conservation. With ongoing innovations, biological research is set to redefine the boundaries of science and technology.
Conclusion
Biological research continues to push the boundaries of human knowledge, shaping the future of healthcare, environmental conservation, and technological innovation. By embracing ethical considerations and leveraging advanced technologies, researchers can unlock new possibilities in science, paving the way for a healthier and more sustainable world.
0 notes
Text
Fwd: Graduate position: Geneva.MyxomycetesSystematicsEvol
Begin forwarded message: > From: [email protected] > Subject: Graduate position: Geneva.MyxomycetesSystematicsEvol > Date: 9 August 2024 at 06:10:32 BST > To: [email protected] > > > The Conservatory and Botanical Garden of Geneva (CJBG) and Geneva > University (UniGE) seek a PhD student > > Title of the project: A biosystematic revision of the family Cribrariaceae > Function: Doctoral candidate and university assistant, 70% > Contract starting date: 1 February 2025 (4 years) > > Job description: > The present project is focused on the systematics and evolution of one of > the most neglected groups of *Myxomycetes* (*Amoebozoa*), the family > *Cribrariaceae* (ca. 50 accepted species). The generic and species > boundaries, as well as the evolution of phenotypic traits, have never been > analysed in detail, and especially not in a phylogenetic context and with > modern techniques. We will assess the phylogenetic relationships of the > species using a fair taxonomic and molecular sampling, including NGS > techniques and phylogenomic analyses. Next, we will explore the evolution > of some phenotypic traits (notably, the sporophore macromorphology and the > spore ornamentation, but also other characters) and the ecology of the > species, to assess its relevance for the taxonomy and diversification of > the group. Finally, we will address the species-level taxonomy, identify > new species and cryptic taxa, and evaluate synonymies. With all this > information, we expect to propose a revised classification and an updated > monograph of the family. Most of the skills and data analyses learned by > the candidate upon completion of the project will also be applicable to the > systematics, taxonomy, and evolutionary biology of other groups of > organisms. > > Within the research project, the candidate will profit from aid, guidance, > and supervision in the different steps of the project, from specimen > handling, data gathering and analyses, to presentation of the results, and > scientific writing. In addition, the candidate is expected to build up a > network of national and international collaborations, perform short stays > abroad, and become used to the dynamic of scientific research to become an > independent researcher. > The assistant position is under the direction of Dr. Juan Carlos Zamora > (CJBG) and Dr. Carlos Lado (RJB-CSIC), and Yamama Naciri (CJBG, UniGE) as > responsible professor. > > Required degrees and skills: > - Master’s degree in biology or in a Biosystematics-related topic in > Environmental/Chemistry/Health Sciences. > - Motivation to engage in the research project and in scientific research, > including: (i) field work and in vitro cultures, (ii) data gathering and > analyses, (iii) writing of a doctoral thesis, (iv) presentation and > dissemination of the project results (scientific journal publication, > congresses, and others). > - Skills in Fungal/Botanical Systematics and Taxonomy, especially > microscopy. > - Skills in molecular lab and phylogenetic analyses. > - Interest for the morphological, evolutionary, and taxonomic study of > Myxomycetes. > - Good disposition to work in a team and to participate in the scientific > life of the host institute. > - Proficiency in written and spoken French (mother tongue or a minimum of > level B2 in CEFR scale) to ensure an adequate flow of the practical classes > of the Systematics and Biodiversity course, given in Biology to 2 year > students, which she/he would be responsible for. > - The working language of the project being English, the candidate must > also be particularly comfortable in this language (written and spoken) > > Application process: > Application deadline: 31 August 2024 > The application file (in English), including (1) a motivation letter and > (2) a detailed CV, should be sent exclusively by e-mail to > *[email protected]* (Juan Carlos Zamora), cc. > *[email protected]*, *[email protected]* > > Juan Carlos Zamora Señoret
0 notes
Text
Aptamers Market Analysis Demand, Statistics, Top Manufacturers, Revenue by Reports and Insights 2030
The latest market report published by Credence Research, Inc. “Global Aptamers Market: Growth, Future Prospects, and Competitive Analysis, 2022 – 2030. The aptamers market is projected to reach sales of USD 858.74 million by 2030, displaying a compound annual boom fee (CAGR) of 17.08% throughout the forecast from 2023 to 2030.
Aptamers, often referred to as "chemical antibodies," are short single-stranded DNA or RNA sequences meticulously crafted through systematic evolution. This process, known as SELEX (Systematic Evolution of Ligands by Exponential Enrichment), empowers scientists to design aptamers with a high degree of specificity and affinity to target molecules. This unique attribute grants aptamers a remarkable edge in diagnostic applications.
Aptamers Market Dynamics refers to the ever-changing landscape of the aptamers market, which is driven by a multitude of factors. These dynamics are characterized by constant fluctuations in demand and supply, technological advancements, regulatory frameworks, and competitive forces. The aptamers market has witnessed significant growth over the years due to its broad range of applications in therapeutics, diagnostics, research, and development. Factors such as increasing investment in research and development activities, growing prevalence of chronic diseases like cancer and diabetes, rising demand for personalized medicine solutions, and advancements in next-generation sequencing technologies have propelled market expansion.
The Versatility Unleashed: Aptamer Applications
1. Clinical Diagnostics
The medical realm is experiencing an undeniable transformation with aptamers taking center stage. These miniature marvels exhibit exceptional binding capabilities, enabling them to identify specific biomarkers indicative of diseases. The implications are profound – early detection, precise prognoses, and tailored treatment strategies. Aptamers redefine the scope of personalized medicine, fostering a new era of patient care.
2. Environmental Monitoring
Beyond healthcare, aptamers extend their prowess to environmental monitoring. By selectively detecting pollutants, contaminants, and hazardous agents, these molecular sentinels contribute to safeguarding our ecosystems. The marriage of aptamers and cutting-edge sensor technologies enhances our ability to address environmental challenges and uphold sustainability.
3. Food Safety Assurance
Aptamers transcend boundaries, ensuring the safety and integrity of our food supply. Rapid and accurate detection of pathogens, allergens, and adulterants is achievable through aptamer-based assays. This advancement not only enhances consumer well-being but also fortifies the food industry's commitment to quality control.
Browse 235 pages report Aptamers Market Global Aptamers Market By Segment ( Nucleic Acid Aptamer, Peptide Aptamer), – By Application ( Diagnostics, Therapeutics, Research & Development) - Growth, Future Prospects & Competitive Analysis, 2016 – 2030)- https://www.credenceresearch.com/report/aptamers-market
Pioneering Research: Illuminating Aptamer Development
Behind every groundbreaking discovery lies pioneering research. In the realm of aptamers, scientists continually refine SELEX methodologies, harnessing computational modeling and innovative selection techniques. The convergence of biology, chemistry, and informatics fuels the evolution of aptamer design, propelling diagnostic capabilities to unprecedented heights.
Advancing Diagnostics: The Road Ahead
The journey of aptamers has just begun, and the road ahead is brimming with possibilities. Collaborative efforts between academia, industry, and healthcare practitioners fuel the translation of aptamer-based innovations into tangible solutions. As aptamers continue to defy limits and expand diagnostic horizons, their role in shaping a healthier, safer, and more sustainable future remains indisputable.
In conclusion, aptamers stand as a testament to human ingenuity, pushing the boundaries of diagnostics with unwavering determination. The convergence of cutting-edge science, technology, and a shared vision propels aptamers into the spotlight of innovation. As we unravel the intricacies of these molecular marvels, we pave the way for a new era of diagnostics that promises to transform lives and industries alike.
Why to Buy This Report-
The report provides a qualitative as well as quantitative analysis of the global Aptamers Market by segments, current trends, drivers, restraints, opportunities, challenges, and market dynamics with the historical period from 2016-2020, the base year- 2021, and the projection period 2022-2028.
The report includes information on the competitive landscape, such as how the market's top competitors operate at the global, regional, and country levels.
Major nations in each region with their import/export statistics
The global Aptamers Market report also includes the analysis of the market at a global, regional, and country-level along with key market trends, major players analysis, market growth strategies, and key application areas.
Browse Full Report: https://www.credenceresearch.com/report/aptamers-market
Visit: https://www.credenceresearch.com/
Related Report: https://www.credenceresearch.com/report/biologic-imaging-reagents-market
Related Report: https://www.credenceresearch.com/report/agriculture-biotechnology-market
Browse our Blog: https://www.linkedin.com/pulse/aptamers-market-analysis-demand-statistics-top-revenue-singh
Browse Our Blog: https://tealfeed.com/aptamers-market-rising-trends-research-outlook-4zylr
About Us -
Credence Research is a viable intelligence and market research platform that provides quantitative B2B research to more than 10,000 clients worldwide and is built on the Give principle. The company is a market research and consulting firm serving governments, non-legislative associations, non-profit organizations, and various organizations worldwide. We help our clients improve their execution in a lasting way and understand their most imperative objectives. For nearly a century, we’ve built a company well-prepared for this task.
Contact Us:
Office No 3 Second Floor, Abhilasha Bhawan, Pinto Park, Gwalior [M.P] 474005 India
0 notes
Text
GATE Life Science Question Paper, Exam Pattern, Mock Test, MCQ
GATE Life Science Question Paper, Exam Pattern, Mock Test, MCQ The Graduate Aptitude Test in Engineering is an examination that primarily tests the comprehensive understanding of various undergraduate subjects in engineering and science for admission into the Masters Program of institutes as well as jobs at Public Sector Companies. GATE Life Science Question Paper and MCQs Buy the question bank or online quiz of GATE Life Science Exam Going through the GATE Life Science Exam Question Bank is a must for aspirants to both understand the exam structure as well as be well prepared to attempt the exam. The first step towards both preparation as well as revision is to practice from GATE Life Science Exam with the help of Question Bank or Online quiz. We will provide you the questions with detailed answer. GATE Life Science Question Paper and MCQs : Available Now GATE Life Science Mock Test Crack GATE Life Science Recruitment exam with the help of online mock test Series or Free Mock Test. Every Sample Paper in GATE Life Science Exam has a designated weightage so do not miss out any Paper. Prepare and Practice Mock for GATE Life Science exam and check your test scores. You can get an experience by doing the Free Online Test or Sample Paper of GATE Life Science Exam. Free Mock Test will help you to analysis your performance in the Examination. GATE Life Science Mock Test : Available Now GATE Life Science Syllabus 1. Chemistry (Compulsory) : Atomic structure and periodicity: Planck’s quantum theory, wave particle duality, uncertainty principle, quantum mechanical model of hydrogen atom, electronic configuration of atoms and ions. Periodic table and periodic properties: ionization energy, electron affinity, electronegativity and atomic size. Structure and bonding: Ionic and covalent bonding, MO and VB approaches for diatomic molecules, VSEPR theory and shape of molecules, hybridization, resonance, dipole moment, structure parameters such as bond length, bond angle and bond energy, hydrogen bonding and van der Waals interactions. Ionic solids, ionic radii and lattice energy (Born‐Haber cycle). HSAB principle. s.p. and d Block Elements: Oxides, halides and hydrides of alkali, alkaline earth metals, B, Al, Si, N, P, and S. General characteristics of 3d elements. Coordination complexes: valence bond and crystal field theory, color, geometry, magnetic properties and isomerism. Chemical Equilibria: Colligative properties of solutions, ionic equilibria in solution, solubility product, common ion effect, hydrolysis of salts, pH, buffer and their applications. Equilibrium constants (Kc, Kp and Kx) for homogeneous reactions. Electrochemistry: Conductance, Kohlrausch law, cell potentials, emf, Nernst equation, Galvanic cells, thermodynamic aspects and their applications. Reaction Kinetics: Rate constant, order of reaction, molecularity, activation energy, zero, first and second order kinetics, catalysis and elementary enzyme reactions. Thermodynamics: First law, reversible and irreversible processes, internal energy, enthalpy, Kirchoff equation, heat of reaction, Hess’s law, heat of formation. Second law, entropy, free energy and work function. Gibbs‐Helmholtz equation, Clausius‐Clapeyron equation, free energy change, equilibrium constant and Trouton’s rule. Third law of thermodynamics Plant Pathology: Nature and classification of plant diseases, diseases of important crops caused by fungi, bacteria,nematodes and viruses, and their control measures, mechanism(s) of pathogenesis and resistance, molecular detection of pathogens; plant-microbe beneficial interactions. Ecology and Environment: Ecosystems – types, dynamics, degradation, ecological succession; food chains and energy flow; vegetation types of the world, pollution and global warming, speciation and extinction, conservation strategies, cryopreservation, phytoremediation. 3. Microbiology Historical Perspective: Discovery of microbial world; Landmark discoveries relevant to the field of microbiology; Controversy over spontaneous generation; Role of microorganisms in transformation of organic matter and in the causation of diseases. Methods in Microbiology: Pure culture techniques; Theory and practice of sterilization; Principles of microbial nutrition; Enrichment culture techniques for isolation of microorganisms; Light-, phase contrast- and electron-microscopy. Microbial Taxonomy and Diversity: Bacteria, Archea and their broad classification; Eukaryotic microbes: Yeasts, molds and protozoa; Viruses and their classification; Molecular approaches to microbial taxonomy. Prokaryotic and Eukaryotic Cells: Structure and Function: Prokaryotic Cells: cell walls, cell membranes, mechanisms of solute transport across membranes, Flagella and Pili, Capsules, Cell inclusions like endospores and gas vesicles; Eukaryotic cell organelles: Endoplasmic reticulum, Golgi apparatus, mitochondria and chloroplasts. Microbial Growth: Definition of growth; Growth curve; Mathematical expression of exponential growth phase; Measurement of growth and growth yields; Synchronous growth; Continuous culture; Effect of environmental factors on growth. Control of Micro-organisms: Effect of physical and chemical agents; Evaluation of effectiveness of antimicrobial agents. Microbial Metabolism: Energetics: redox reactions and electron carriers; An overview of metabolism; Glycolysis; Pentose-phosphate pathway; Entner-Doudoroff pathway; Glyoxalate pathway; The citric acid cycle; Fermentation; Aerobic and anaerobic respiration; Chemolithotrophy; Photosynthesis; Calvin cycle; Biosynthetic pathway for fatty acids synthesis; Common regulatory mechanisms in synthesis of amino acids; Regulation of major metabolic pathways. Microbial Diseases and Host Pathogen Interaction: Normal microbiota; Classification of infectious diseases; Reservoirs of infection; Nosocomial infection; Emerging infectious diseases; Mechanism of microbial pathogenicity; Nonspecific defense of host; Antigens and antibodies; Humoral and cell mediated immunity; Vaccines; Immune deficiency; Human diseases caused by viruses, bacteria, and pathogenic fungi. Chemotherapy/Antibiotics: General characteristics of antimicrobial drugs; Antibiotics: Classification, mode of action and resistance; Antifungal and antiviral drugs. Microbial Genetics: Types of mutation; UV and chemical mutagens; Selection of mutants; Ames test for mutagenesis; Bacterial genetic system: transformation, conjugation, transduction, recombination, plasmids, transposons; DNA repair; Regulation of gene expression: repression and induction; Operon model; Bacterial genome with special reference to E.coli; Phage λ and its life cycle; RNA phages; RNA viruses; Retroviruses; Basic concept of microbial genomics. Microbial Ecology: Microbial interactions; Carbon, sulphur and nitrogen cycles; Soil microorganisms associated with vascular plants. 4. Zoology Animal world: Animal diversity, distribution, systematics and classification of animals, phylogenetic relationships. Evolution: Origin and history of life on earth, theories of evolution, natural selection, adaptation, speciation. Genetics: Basic Principles of inheritance, molecular basis of heredity, sex determination and sex-linked characteristics, cytoplasmic inheritance, linkage, recombination and mapping of genes in eukaryotes, population genetics. Biochemistry and Molecular Biology: Nucleic acids, proteins, lipids and carbohydrates; replication, transcription and translation; regulation of gene expression, organization of genome, Kreb’s cycle, glycolysis, enzyme catalysis, hormones and their actions, vitamins Cell Biology: Structure of cell, cellular organelles and their structure and function, cell cycle, cell division, chromosomes and chromatin structure. Gene expression in Eukaryotes : Eukaryotic gene organization and expression (Basic principles of signal transduction). Animal Anatomy and Physiology: Comparative physiology, the respiratory system, circulatory system, digestive system, the nervous system, the excretory system, the endocrine system, the reproductive system, the skeletal system, osmoregulation. Parasitology and Immunology: Nature of parasite, host-parasite relation, protozoan and helminthic parasites, the immune response, cellular and humoral immune response, evolution of the immune system. Development Biology: Embryonic development, cellular differentiation, organogenesis, metamorphosis, genetic basis of development, stem cells. Ecology: The ecosystem, habitats, the food chain, population dynamics, species diversity, zoogerography, biogeochemical cycles, conservation biology. Animal Behaviour: Types of behaviours, courtship, mating and territoriality, instinct, learning and memory, social behaviour across the animal taxa, communication, pheromones, evolution of animal behaviour. GATE 2021 LX Exam Pattern Duration : 180 Mint Negative Mark : 0.66 SectionNo. of QuestionsMarksMarks/QuestionsTotal Marks General Aptitude5 55 51 25 10 Technical, Engineering, Mathematics25 3025 301 225 60 Total65 100
2 notes
·
View notes
Photo



The lost art of looking at plants
When Elizabeth Kellogg finished her PhD in 1983, she feared that her skills were already obsolete. Kellogg studied plant morphology and systematics: scrutinizing the dazzling variety of plants’ physical forms to tease out how different species are related. But most of her colleagues had already pivoted to a new approach: molecular biology. “Every job suddenly required molecular techniques,” she says. “It was like I had learned how to make illuminated manuscripts, and then somebody invented the printing press.”
Kellogg had graduated near the start of a revolution in plant biology. Over the next few decades, as researchers adopted molecular tools and DNA sequencing, detailed analyses of plants’ physical traits fell out of fashion. And because many geneticists worked with only a few key organisms, such as the thale cress Arabidopsis thaliana, they didn’t need expertise in comparing and contrasting different plant species. At universities, botany departments folded and molecular-biology departments swelled. Kellogg, now at the Donald Danforth Plant Science Center in St Louis, Missouri, adapted: she embraced genomics, and combined it with her morphology skills to trace the evolution of key traits in the wild relatives of food crops.
But lately, Kellogg has noticed a resurgence of interest in the old ways. Advances in imaging technology — allowing researchers to peer inside plant structures in 3D — mean that biologists are seeking expertise in plant physiology and morphology again. And improvements in gene editing and sequencing have liberated geneticists to tinker with DNA in a wider range of flora, giving them a renewed appetite to understand plant diversity.
Plant biologists hope that, by combining new approaches to botany with data from genomics and imaging labs, they can provide better answers to questions that biologists have asked for more than 100 years: how genes and the environment shape the rich diversity of plants’ physical forms. “People are starting to look beyond their own system into plants as a whole,” says Kellogg. Plant morphology was once a science of form for its own sake, she says, but now, it is being pressed into service to understand how plant traits connect to gene activity across disparate species. “It’s coming back — just under different guises.”
3D imaging offers new views of these antirrhinum (snapdragon) buds. Credit: Karen Lee/Xana Rebocho/John Innes Centre
Pollen masses inside a 'deceptive' orchid (Orchis militaris), imaged using X-ray computed tomography.Credit: Yannick Staedler
Watching leaves grow: individual cells in this 7-day-old Cardamine hirsuta mustard leaf are outlined by labelling cell membranes with fluorescent molecules.Credit: Miltos Tsiantis/Daniel Kierzkowski
270 notes
·
View notes
Link
When Elizabeth Kellogg finished her PhD in 1983, she feared that her skills were already obsolete. Kellogg studied plant morphology and systematics: scrutinizing the dazzling variety of plants’ physical forms to tease out how different species are related. But most of her colleagues had already pivoted to a new approach: molecular biology. “Every job suddenly required molecular techniques,” she says. “It was like I had learned how to make illuminated manuscripts, and then somebody invented the printing press.”
Kellogg had graduated near the start of a revolution in plant biology. Over the next few decades, as researchers adopted molecular tools and DNA sequencing, detailed analyses of plants’ physical traits fell out of fashion. And because many geneticists worked with only a few key organisms, such as the thale cress Arabidopsis thaliana, they didn’t need expertise in comparing and contrasting different plant species. At universities, botany departments folded and molecular-biology departments swelled. Kellogg, now at the Donald Danforth Plant Science Center in St Louis, Missouri, adapted: she embraced genomics, and combined it with her morphology skills to trace the evolution of key traits in the wild relatives of food crops.
But lately, Kellogg has noticed a resurgence of interest in the old ways. Advances in imaging technology — allowing researchers to peer inside plant structures in 3D — mean that biologists are seeking expertise in plant physiology and morphology again. And improvements in gene editing and sequencing have liberated geneticists to tinker with DNA in a wider range of flora, giving them a renewed appetite to understand plant diversity.
124 notes
·
View notes
Link
If We Made Life in a Lab, Would We Understand It Differently?
Rebecca Wilbanks
Aug 19, 2018
What is life? For much of the 20th century, this question did not particularly concern biologists. Life is a term for poets, not scientists, argued the synthetic biologist Andrew Ellington in 2008, who began his career studying how life began. Despite Ellington’s reservations, the related fields of origins-of-life research and astrobiology have renewed focus on the meaning of life. To recognize the different form that life might have taken four billion years ago, or the shape it could take on other planets, researchers need to understand what, in essence, makes something alive.
Life, however, is a moving target, as philosophers have long observed. Aristotle distinguished “life” as a concept from “the living”—the collection of existing beings that make up our world, such as the neighbor’s dog, my cousin and the bacteria growing in your sink. To know life, we must study the living; but the living is always changing across time and space. In trying to define life, we must consider the life we know and the life we don’t know. As the origins-of-life researcher Pier Luigi Luisi at Roma Tre University puts it, there is life-as-it-is-now, life-as-it-could-be, and life-as-it-once-was. These categories point to a dilemma that medieval mystical philosophers addressed. Life, they noticed, is always more than the living, making it, paradoxically, permanently inaccessible to the living. Because of this gap between actual life and potential life, many definitions of life focus on its capacity to change and evolve rather than trying to pin down fixed characteristics.
In the early 1990s while advising NASA on the possibilities of life on other planets, the biologist Gerald Joyce, now at the Salk Institute for Biological Studies in California, helped to come up with one of the most widely used definitions of life. It’s known as the chemical Darwinian definition: “Life is a self-sustained chemical system capable of undergoing Darwinian evolution.” In 2009, after decades of work, Joyce’s group published a paper in which they described an RNA molecule that could catalyze its own synthesis reaction to make more copies of itself. This chemical system met Joyce’s definition of life. But nobody wanted to claim that it was alive. The problem was, it hadn’t done anything new or exciting yet. A New York Times article put it this way: “Someday their genome may surprise their creator with a word—a trick or a new move in the game of almost life—that he has not anticipated. ‘If it would happen, if it would do it for me, I would be happy,’ Dr Joyce said, adding, ‘I won’t say it out loud, but it’s alive.’ ”
Joyce seeks to understand life by trying to generate simple living systems in the lab. In doing so, he and other synthetic biologists bring new kinds of life into being. Every attempt to synthesize novel life forms points to the fact that there are still more, perhaps infinite, possibilities for how life could be. Synthetic biologists could change the way life evolves, or its capacity to evolve at all. Their work raises new questions about a definition of life based on evolution. How to categorize life that is redesigned, the product of a break in the chain of evolutionary descent?
An origin story for synthetic biology goes like this: in 1997, Drew Endy, one of the founders of synthetic biology and now a professor of bioengineering at Stanford University in California, was trying to create a computational model of the simplest life form he could find: the bacteriophage T7, a virus that infects E coli bacteria. A crystalline head atop spindly legs, it looks like a landing capsule touching down on the Moon as it grabs onto its bacterial host. The bacteriophage is so simple that by some definitions it is not even alive. (Like all viruses, it depends on the molecular machinery of its host cell to replicate.) Bacteriophage T7 has only 56 genes, and Endy thought it might be possible to create a model that accounted for every part of the phage and how those parts worked together: a perfect representation that would predict how the phage would change if any one of its genes were moved or deleted.
Endy built a series of bacteriophage T7 mutants, systematically knocking out genes or scrambling their location in the tiny T7 genome. But the mutant phages conformed to the model only some of the time. A change that should have caused them to weaken would instead have their progeny bursting open E coli cells twice as fast as before. It wasn’t working. Eventually, Endy had a realization: “If we want to model the natural world, we have to rewrite [the natural world] to be modellable.” Instead of trying to make a better map, change the territory. Thus was born the field of synthetic biology. Borrowing techniques from software engineering, Endy began to “refactor” bacteriophage T7’s genome. He made bacteriophage T7.1, a life form designed for ease of interpretation to the human mind.
Phage T7.1 is an example of what one synthetic biologist has called supra-Darwinian life: life that owes its existence to human design, rather than natural selection. Bioengineers such as Endy approach life in dualistic terms: a physical structure on the one hand, a pattern of information on the other. In theory, a perfect representation of life would enable a seamless transition between information and matter, intention and realization: change some letters of DNA on your computer screen, print out an organism that looks and behaves just as you intended. With this approach, evolution threatens to corrupt the engineer’s blueprint. Preserving one’s biological designs might require making your engineered organisms unable to reproduce or evolve.
In contrast, Joyce’s desire for his molecules to surprise him suggests that the capacity for open-ended evolution — “inventiveness, pluripotentiality, open-endedness” — is the critical criteria of life. In accordance with this idea, Joyce now defines life as “a genetic system that contains more bits [of information] than the number that were required to initiate its operation.” But according to this definition, given two identical systems with different histories—one designed and the other evolved—only the latter would be considered alive; the rationally designed system, no matter how complex, would be just a “technological artifact.”
Design and evolution are not always opposed. Many synthetic biology projects use a mix of rational design and directed evolution: they construct a host of mutant cells—variations on a theme—and select the ones that work the best. Although Joyce’s new understanding of life still involves evolution, it evokes the abrupt temporality of emergence rather than Darwin’s longue durée. Emergent life fits a culture of disruptive innovation whose ultimate ideal approximates something like the magic of pulling a kidney out of a 3D printer: the enchantment of joining together familiar things with new and surprising results. Design and evolution are also compatible when bioengineers look at genetic diversity as a treasure trove of design elements for future life forms.
For some synthetic biologists, the path to what the mystics called life-beyond-life—life that exceeds the living as we know it—now runs through biological engineering. Endy describes his vocation in terms of a desire to contribute to life by generating new kinds of “improbable patterns that continue to thrive and exist.” Joyce imagines life and technology joining forces against the fundamental thermodynamic tendency towards disorder and decay. What new forms life will take, only time will tell.
This article was originally published at Aeon and has been republished under Creative Commons.
Image Credit: nobeastsofierce / Shutterstock.com
1 note
·
View note
Text
Aptamers Market 2020 Industry Overview, Trends, Growing Demand, Growth, Production, Types, Applications and Forecast to 2027
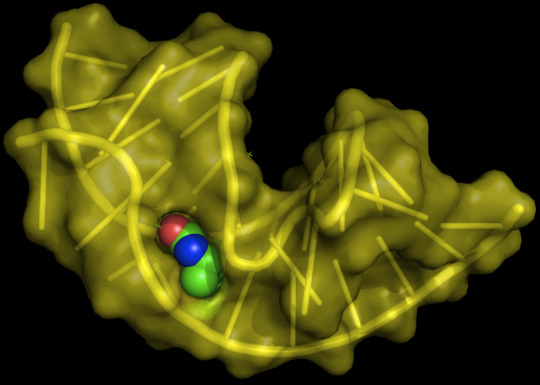
Aptamers are short single-stranded DNA or RNA (ssDNA or ssRNA) molecules that bind to a specific target protein, lipid, or nucleic acid molecules with high specificity. Aptamers can also be conjugated to drugs, and work as drug-delivery systems. For example, aptamers have been conjugated with chemotherapy drugs to turn target and bind to only cancerous cells and leave healthy tissue untouched.
Furthermore, factors such as lesser possibility of provoking undesired immune responses, high thermal stability, and cost-efficiency provide are expected to boost demand for aptamers. These are produced through a technique called Systematic Evolution of Ligands by Exponential enrichment (SELEX), which is time-consuming and involves iterative steps. Thus, to overcome this disadvantage, Vivonics Inc. developed and patented a one-step rapid technique named Rapid Isolation of DNA Aptamers (RIDA) for the isolation of high affinity and high selectivity aptamers. Such factors are expected to fuel adoption of aptamers.
The global aptamers market size was valued at US$ 3,626.13 million in 2019, and is expected to witness a CAGR of 18.5% over the forecast period (2019 – 2027).
Request for Sample Copy: https://www.coherentmarketinsights.com/insight/request-sample/774
Aptamers have attracted much attention recently for potential therapeutic and diagnostic purposes and have gained worldwide preference over antibodies owing to the former’s key benefits – lower cost, ability to not destroy neighboring cells, greater binding affinity, longer shelf life and, high specificity. The wide range of applications of aptamers has dominate medical industry to convey monetary clinical evidence and allow companies to significantly increase their influence to capture the untapped potential of aptamers, functioning as a key development opportunity that will propel the market growth.
An increasing number of applications is expected to drive the market growth
Aptamer products can be used as biosensors, diagnostics research agents or can be considered as a tool for biomarker or drug discovery. It can also be used for targeted therapeutics and bio-industrial applications. In-vivo aptamers can avoid majority of bench testing, save time, and around 35% of the R&D cost. It permits the natural selection of drug candidates in whole animal models, which reduces the false starts and requires fewer animals for drug evaluation. All these factors have led a large number of companies to invest in this market.
Moreover, growing incidence of life-threatening diseases such as cancer has directed many companies to introduce new detection assays based on aptamers therapy which is expected to help drive the growth of the aptamers market. In March 2017, Aptamer Sciences, Inc. unveiled the multi-protein biomarker assay by using this technology for detection of lung cancer.
Global Aptamers Market – Restraints
Stringent regulations related the use of aptamers and low awareness about these molecules in low- and middle-income countries are some of the factors that may challenge the growth of the market.
Global Aptamers Market - Regional Insights
On the basis of region, the global aptamers market is segmented into North America, Latin America, Europe, Asia Pacific, Middle East, and Africa. North America holds a dominant position in the global aptamers market, owing to presence of major key players and adoption of strategic collaborations. For instance, in March 2014, Siva Therapeutics, a biotechnology company initiated a research collaboration with SomaLogic, Inc., clinical diagnostic company to determine the ability of SomaLogic’s SOMAmer (Slow-off Rate Modified Aptamer) reagents to actively target gold nanorods to solid tumors. This is expected to create more applications of aptamers in the near future.
Asia Pacific is the fastest growing region in the global aptamers market owing to increasing research and development in the field of aptamers. For instance, in 2015, scientists from the National Institute of Pharmaceutical Research, Mohali India, described the molecular mechanism of RNA aptamers which specifically targets drug resistant and metastatic cancer cells. The NIPER researchers selected the RNA aptamer through Cell-SELEX process.
To Get Instant Access, Purchase Report Here @ https://www.coherentmarketinsights.com/insight/buy-now/774
Global Aptamers Market - Competitive Landscape
Key players operating in the global aptamers market include Ophthotech Corporation, Aptamer Sciences, Inc., Novartis AG, SomaLogic, Inc., NeoNeuro SAS, Aptagen LLC, Aptus Biotech S.L., AM Biotechnologies, LLC, Base Pair Biotechnologies and Vivonics Inc.
About Coherent Market Insights
Coherent Market Insights is a prominent market research and consulting firm offering action-ready syndicated research reports, custom market analysis, consulting services, and competitive analysis through various recommendations related to emerging market trends, technologies, and potential absolute dollar opportunity.
Contact Us
Mr. Shah
Coherent Market Insights
1001 4th Ave, #3200
Seattle, WA 98154
Phone: US +12067016702 / UK +4402081334027
Email: [email protected]
0 notes
Link
In The Invertebrate Tree of Life, Gonzalo Giribet and Gregory Edgecombe, leading authorities on invertebrate biology and paleontology, utilize phylogenetics to trace the evolution of animals from their origins in the Proterozoic to today. Phylogenetic relationships between and within the major animal groups are based on the latest molecular analyses, which are increasingly genomic in scale and draw on the soundest methods of tree reconstruction. Giribet and Edgecombe evaluate the evolution of animal organ systems, exploring how current debates about phylogenetic relationships affect the ways in which aspects of invertebrate nervous systems, reproductive biology, and other key features are inferred to have developed. The authors review the systematics, natural history, anatomy, development, and fossil records of all major animal groups, employing seminal historical works and cutting-edge research in evolutionary developmental biology, genomics, and advanced imaging techniques. Overall, they provide a synthetic treatment of all animal phyla and discuss their relationships via an integrative approach to invertebrate systematics, anatomy, paleontology, and genomics. With numerous detailed illustrations and phylogenetic trees, The Invertebrate Tree of Life is a must-have reference for biologists and anyone interested in invertebrates, and will be an ideal text for courses in invertebrate biology.


0 notes
Text
Why viruses like Herpes and Zika will need to be reclassified, and its biotech impact

New research reveals that the way viruses were perceived in terms of their architecture will need to be retooled, because they are actually structured in many more patterns than previously understood. The findings could have significant impact on how they are classified, our understanding of how they form, evolve and infect hosts, and strategies to identify ways to design vaccines to target them. In the 1950s and '60s as scientists began to obtain high resolution images of viruses, they discovered the detailed structure of the capsid—an outer protective layer composed of multiple copies of the same protein—which protects the virus' genetic material. The majority of viruses have capsids that are typically quasi-spherical and display icosahedral symmetry—like a 20-sided dice for instance. The capsid shell is what protects them, and as scientists discovered their structure, they proposed that capsids could have different sizes and hold different amounts of genome, and therefore could infect hosts differently. Why this matters When designing drugs to target viruses, scientists can now take their varying structural shapes into account to improve efficacy. Two researchers who study the structures of viruses, Antoni Luque, a theoretical biophysicist at San Diego State University and a member of its Viral Information Institute, and Reidun Twarock, a mathematical biologist from the University of York, UK, and a member of York's Cross-disciplinary Centre for Systems Analysis, show that many viruses have essentially been misclassified for 60 years, including common viruses such as Herpes simplex and Zika. This was because despite having the structural images from cryo-electron microscopy, we did not have the mathematical description of many of the architectures of different viruses. "We discovered six new ways in which proteins can organize to form icosahedral capsid shells," Luque said. "So, many viruses don't adopt only the two broadly understood capsid architectures. There are now at least eight ways in which their icosahedral capsids could be designed." They used a generalization of the quasiequivalence principle to see how proteins can wrap around an icosahedral capsid. Their study, which will be published in Nature Communications on Friday, September 27, also shows that viruses that are part of the same structural lineage, based on the protein that they're composed of, adopt consistent icosahedral capsid layouts, providing a new approach to study virus evolution. Biotech applications Structural biologists can now take this information and reclassify the structure of the viruses, which will help unveil molecular and evolutionary relationships between different viruses. It will also provide a guide to engineer new molecular containers for nanotech and biotech applications, and it will help scientists to identify specific strategies to target the assembly of proteins in the capsid. This can eventually lead to a more systematic approach to developing antiviral vaccines. "We can use this discovery to target both the assembly and stability of the capsid, to either prevent the formation of the virus when it infects the host cell, or break it apart after it's formed," Luque said. "This could facilitate the characterization and identification of antiviral targets for viruses sharing the same icosahedral layout." This new framework accommodates viruses that were previously outliers, provides new predictions of viral capsid architectures, and has identified common geometrical patterns among distant evolutionary related viruses that infect everyone from humans to bacteria. Twarock said the new blueprints also provide "a new perspective on viral evolution, suggesting novel routes in which larger and more complex viruses may have evolved from simple ones at evolutionary timescales." Architectural applications The geometries could be also used in new architectural designs in buildings and construction. Since the 1960s, these viral capsids have been classified using the geometrical framework introduced by structural biologist Donald Caspar and biophysicist Aaron Klug, which were inspired by the geodesic domes designed by the renowned architect R. Buckminster Fuller. However, as molecular imaging techniques have advanced, an increasing number of 3-D viral capsid reconstructions that included viruses like Herpes or Zika have fallen out from this classical geometrical framework. "This study introduces a more general framework than the classic Caspar-Klug construction. It is based on the conservation of the local vertices formed by the proteins that interact in the capsid," Luque explained. "This approach led to the discovery of six new types of icosahedral capsid layouts, while recovering the two classical layouts from Caspar-Klug based on Goldberg and geodesic polyhedra." "Structural puzzles in virology solved with an overarching icosahedral design principle" is published in Nature Communications. Provided by: San Diego State University More information: Reidun Twarock et al. Structural puzzles in virology solved with an overarching icosahedral design principle. Nature Communications (2019). doi.org/10.1038/s41467-019-12367-3 Image: Evolutionary related viruses infecting bacteria and humans adopting one of the newly established protein layouts of icosahedral capsids. Bacillus phage Basilisk (a), herpes simplex virus 1 (b), and bacteriophage lambda (c). Credit: Antoni Luque, San Diego State University and Reidun Twarock, University of York. Read the full article
0 notes
Text
Job: KewGardens.ResAssist.FungiPlantInteractions
Research Assistant in Fungi and Plant Interactions Royal Botanic Gardens, Kew You will join Kew’s science staff as a research assistant, conducting lab and field work and contributing towards publishing oustanding research within the Fungus-Plant Interactions programme. You will be part of the Comparative Fungal Biology team (part of the Comparative Plant and Fungal Biology department) that works closely with the other mycology teams at Kew to research the systematics, ecology and evolution of fungi and their interactions with other organisms. Research ranges from baseline diversity studies in biodiversity hotspots through to reconstructing the Fungal Tree of Life, with a special emphasis on evolution and ecology of lifestyles and symbiotic interactions. You will be working with different types of fungi, but with a particular focus on mycorrhizal and endophytic fungi. You will conduct largely independent work in the lab giving support to different projects, including microscopy, culturing, and various molecular techniques. It is also expected a basic knowledge on statistical analyses and notions of phylogenetics and phylogenomics. You will be an outstanding early career scientist with a BSc or MSc in a relevant subject area. You will have a proven aptitude for delivering scientific outputs and excellent writing and oral skills. You will be an enthusiastic team player who is ready to engage in several projects and work in collaboration with multiple researchers at Kew. Kew is the world’s leading botanic gardens, at the forefront of plant and fungal science, a UNESCO World Heritage Site and a major visitor attraction. Kew’s mission is to inspire a world where plants and fungi are understood, valued and conserved – because our lives depend on them. We use the power of our science and the rich diversity of our gardens and collections to provide knowledge, inspiration and understanding of why plants and fungi matter to everyone. This research assistant position is offered for two years. Salary will be in the range of £24,455 to £27,325 per annum pro rata, depending on skills and experience. Closing Date: 10/03/2019 Further information: http://bit.ly/2STd4M5 We are committed to equality of opportunity and welcome applications from all sections of the community. We guarantee to interview all disabled applicants who meet the essential criteria for the post. Ester Gaya Dr. Ester Gaya Senior Research Leader | Comparative Fungal Biology Jodrell laboratory Royal Botanic Gardens, Kew, Richmond, Surrey, TW9 3DS, UK Tel.:+44(0)208 332 5381 Fax:+44(0)208 332 5310 http://bit.ly/2p9oBG3 Email: [email protected]/ [email protected] The Royal Botanic Gardens, Kew is a non-departmental public body with exempt charitable status, whose principal place of business is at Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3AE, United Kingdom. Ester Gaya via Gmail
1 note
·
View note
Text
Manufacturing Engineering Challenges of Pharmaceutical 3D Printing for on-Demand Drug Delivery - Juniper Publishers
Juniper Publishers - Open Access Journal of Engineering Technology

Abstract
There are about two dozen 3D printing technologies available in the market. These technologies differ in terms of layering mechanism, type of material that can be handled, binding mechanism and suitability for solid dosage form manufacturing. Based on literature information and preliminary data, fused deposition modeling (FDM), selective laser sintering (SLS) and powder-bed 3D printing methods have emerged as the most promising candidates for delivering on-demand drugs according to established Good Manufacturing Practice (GMP) guidelines of the pharmaceutical industry. However, information on interplay of critical process parameters (CPPs) and critical materials attributes (CMAs) on the critical quality attributes (CQAs) of the formulations and products that will determine their in-vitro and in-vivo performance is not readily available in the literature. Through this mini-review, we underline the pressing need for the systematic exploration of these critical factors and present the Ishikawa diagrams for the three mainstream 3D printing techniques already penetrating the specialized niches of the pharmaceutical manufacturing market at an extremely fast pace.
Keywords: Additive Manufacturing; Critical quality attributes; Compatibility; Fused filament fabrication; Micro particles; Nan particles; Ingredients
Abbrevations: AM: Additive Manufacturing; FDM: Fused Deposition Modeling; FFF: Fused Filament Fabrication; SLS: Selective Laser Sintering; PVA: Polyvinyl Alcohol; CPPs: Critical Process Parameters; CQAs: Critical Quality Attributes; CMAs: Critical Material Attributes; GMPs: Good Manufacturing Practices
Introduction
Additive Manufacturing (AM) in general and three-dimensional printing (3DP) in particular is emerging technologies expected to revolutionize pharmaceuticals manufacturing along with other fields [1-4]. These technologies offer the ability to create limitless dosage forms that are likely to challenge conventional drug fabrication methods not only in product quality and efficacy but also in cost efficiency as 3D printers have already been successful in producing novel dosage forms within minutes [3]. Three situations where this on-demand pharmacy capability may be applicable include printing directly on the implants or tissue scaffolds, printing “just in-time” in healthcare facilities or in other resource-constrained settings and printing low-stability drugs for immediate consumption. In all these scenarios, 3D printing technologies provide attractive solutions to explore on-demand pharmacy [5]. FDA approved first commercial drug product based on 3DP technology in 2015 (NDA #207958, Spritam® (levetiracetam) triturates Aprecia Pharmaceuticals Co) [6]. It is a high drug loaded tablet that disintegrates in less than 10 seconds which is atypical even for orally disintegrating tablet. Even though its underlying manufacturing technology is two decades old, very little information is available in the public domain about the process and formulation variables that could affect critical quality attributes of drug products manufactured by 3DP. Furthermore, quality defects in the drug products manufactured by 3DP will be entirely different from compressed tablets. Thus it is essential to evaluate 3DP techniques for their compatibility with pharmaceuticals from the perspective of on-demand pharmacy feasibility and application.
3DP Technologies Amenable to Pharmaceutical Manufacturing and Their Unique Aspects and Materials
The primary 3DP technologies that can be used for pharmaceuticals manufacturing are inkjet-based or inkjet powder-based 3DP, fused deposition modeling (FDM) also called fused filament fabrication (FFF) and selective laser sintering (SLS).
Inkjet-based or inkjet powder-based 3DP
Whether another material or a powder is used as the substrate is what differentiates 3D inkjet printing from powderbased 3D inkjet printing. In inkjet-based drug fabrication, inkjet printers are used to spray formulations of medications and binders in small droplets at precise speeds, motions, and sizes onto a substrate. The most commonly used substrates include different types of cellulose, coated or uncoated paper, microporous bioceramics, glass scaffolds, metal alloys, and potato starch films, among others [2-5]. Researchers have further improved this technology by spraying uniform “ink” droplets onto a liquid film that encapsulates it, forming microparticles and nanoparticles. Such matrices can be used to deliver small hydrophobic molecules and growth factors [7]. In powder-based 3D printing drug fabrication, the inkjet printer head sprays the “ink” onto the powder foundation. When the ink contacts the powder, it hardens and creates a solid dosage form, layer by layer. The ink can include active ingredients as well as binders and other inactive ingredients. After the 3D-printed dosage form is dry, the solid object is removed from the surrounding loose powder substrate [3-5,7]. Very limited work has been reported concerning controllable printing parameters in binder jetting process and materials attributes (active and inactive), which could substantially affect critical quality attributes (CQAs) of pharmaceutical formulations. Therefore, having a good understanding and experimental insight into the practical effects of such parameters on CQAs seems to be essential. The fishbone (Ishikawa) diagram for pharmaceutical powder-bed and inkjet 3D printing processes is given in Figure 1.
Fused deposition modeling (FDM)
In FDM/FFF, the object is formed by layers of melted or softened thermoplastic filament extruded from the printer’s head at specific directions as dictated by computer software. Inside the FDM printer’s head the filament is heated to just above its softening point which is then extruded through a nozzle, deposited layer by layer immediately followed by solidification [4-5,7-9]. The speed of the extruder head may also be controlled to stop and start deposition and form an interrupted plane without stringing or dribbling between sections although nanoparticle emissions were detected during the process [2-3,9-12]. The potential of FDM 3D printing to incorporate drugs into commercially available filaments has been explored previously [13,14]. Nonetheless, all those studies highlighted several challenges involved in employing printing technique for pharmaceutical applications. The use of elevated temperatures (185-220 °C) and limited drug loading (0.063-9.5%w/w) renders it less suitable for many drugs particularly thermolabile ones [13-15]. FDM 3D printing has been also restricted to a number of biodegradable thermoplastic polymers such as polylactic acid (PLA) [16] and polyvinyl alcohol (PVA) [4,13- 15] in comparison to a wide variety of choices for conventional tableting. Despite the attractive properties of PVA [17] and PLA [18], the use of high polymer ratio in combination with the need for high molecular weight of the polymers generates polymeric matrices with limited porosity, thus resulting in extended drug release patterns. To increase the flexibility and potential of FDM 3D printing process, never filament formulations also employing nanocomposite compounding approaches will be needed to overcome existing limitations of FDM type pharmaceutical 3D printing. The fishbone (Ishikawa) diagram for pharmaceutical FDM processes is provided in Figure 2.
Selective laser sintering (SLS)
Like all methods of 3D printing, an object printed with the SLS process starts as a CAD file. Objects printed through this method are made with powder materials, most commonly plastics, such as nylon, which are dispersed in a thin layer on top of the build platform inside an SLS machine. The laser heats the powder either to just below its melting point (sintering) or above its melting point (melt consolidation), which fuses the particles in the powder together into a solid form. Once the initial layer is formed, the platform of the SLS machine drops usually by less than 0.1 mm, thus exposing a new layer of powder for the laser to trace and fuse together. This process continues again and again until the entire object has been printed [2,19]. Very limited information is available in the literature for pharmaceutical application of the SLS technique [5,20]. The characteristic limitations of this process are attributed to incompatibilities between laser energy settings and properties of thermoplastic polymer used to print drug product. It is important to understand the interplay of the SLS critical process parameters (CPPs) and polymeric powder critical material attributes (CMAs) on critical CQAs, as shown by the Ishikawa diagram in Figure 3.
Conclusion
The most promising 3DP technologies for pharmaceutical applications and their manufacturing engineering related challenges were discussed. In all methods, understanding critical process parameters (CPPs) is equally important as understanding critical material attributes (CMAs) to ensure consistent quality of 3D printed solid dosage form. Rigorous experimental studies are needed to validate process and materials engineering correlations between CMAs and CPPs, which can also identify optimal control strategies and potential risk factors for process failure or deficiencies in terms of pharmaceutical good manufacturing practices (GMPs). Including all variants and hybrid approaches, about two dozen 3DP technologies currently exist in the market each with varying capabilities and complexities. Although objective assessments of current process limitations and capabilities assign a higher chance to powder bed and SLS techniques in the race for on-demand drug delivery, the Darwinian dynamics of technology evolution coupled with process engineering and materials technology innovations will determine which particular ones would be indeed the fittest to survive in the domain of pharmaceutical manufacturing.
For more Open Access Journals in Juniper Publishers please click on: https://juniperpublishers.com
For more articles in Open Access Journal of Engineering Technology please click on: https://juniperpublishers.com/etoaj/index.php
For more Open Access Journals please click on: https://juniperpublishers.com
#Engineering Technology open access journals#Juniper Journals Reviews#Juniper publisher journals#Juniper publishers#Open Access Journals#Peer Review Journals
0 notes
Text
Biomolecules, Vol. 8, Pages 83: Molecular Modeling Applied to Nucleic Acid-Based Molecule Development
Molecular modeling by means of docking and molecular dynamics (MD) has become an integral part of early drug discovery projects, enabling the screening and enrichment of large libraries of small molecules. In the past decades, special emphasis was drawn to nucleic acid (NA)-based molecules in the fields of therapy, diagnosis, and drug delivery. Research has increased dramatically with the advent of the SELEX (systematic #evolution of ligands by exponential enrichment) technique, which results in single-stranded DNA or #RNA sequences that bind with high affinity and specificity to their targets. Herein, we discuss the role and contribution of docking and MD to the development and optimization of new nucleic acid-based molecules. This review focuses on the different approaches currently available for molecular modeling applied to NA interaction with proteins. We discuss topics ranging from structure prediction to docking and MD, highlighting their main advantages and limitations and the influence of flexibility on their calculations. http://bit.ly/2PIM6Bw #MDPI
0 notes
Text
Download Test Bank for Biology 7th edition by Campbell and Reece
This is completed downloadable Test Bank for Biology 7th edition by Neil Campbell and Jane Reece
Instant Download Test Bank for Biology 7th edition by Neil Campbell and Jane Reece
View sample:
https://digitalcontentmarket.org/wp-content/uploads/2017/10/Test-Bank-for-Biology-7th-edition-by-Neil-Campbell-and-Jane-Reece.pdf
Product Description:
Neil Campbell and Jane Reece's BIOLOGY remains unsurpassed as the most successful majors biology textbook in the world. This text has invited more than 4 million students into the study of this dynamic and essential discipline.The authors have restructured each chapter around a conceptual framework of five or six big ideas. An Overview draws students in and sets the stage for the rest of the chapter, each numbered Concept Head announces the beginning of a new concept, and Concept Check questions at the end of each chapter encourage students to assess their mastery of a given concept. New Inquiry Figures focus students on the experimental process, and new Research Method Figures illustrate important techniques in biology. Each chapter ends with a Scientific Inquiry Question that asks students to apply scientific investigation skills to the content of the chapter.
Table of Contents:
Chapter 1 - Exploring Life
Chapter 2 - The Chemical Context of Life
Chapter 3 - Water and the Fitness of the Environment
Chapter 4 - Carbon and the Molecular Diversity of Life
Chapter 5 - The Structure and Function of Macromolecules
Chapter 7 - Membrane Structure and Function
Chapter 8 - An Introduction to Metabolism
Chapter 9 - Cellular Respiration: Harvesting Chemical Energy
Chapter 10 - Photosynthesis
Chapter 11- Cell Communication
Chapter 12 - The Cell Cycle
Chapter 13 - Meiosis and Sexual
Chapter 14 - Mendel and the Gene Idea
Chapter 15 - The Chromosomal Basis of Inheritance
Chapter 16 - The Molecular Basis of Inheritance
Chapter 17 - From Gene to Protein
Chapter 18 - The Genetics of Viruses and Bacteria
Chapter 19 - Eukaryotic Genomes: Organization, Regulation, and Evolution
Chapter 20 - DNA Technology
Chapter 21 - The Genetic Basis
Chapter 22 - Descent with Modification: A Darwinian View of Life
Chapter 23 - The Evolution of Populations
Chapter 24 - The Origin of Species
Chapter 25 - Phylogeny and Systematics
Chapter 26 - The Tree of Life
Chapter 27 - Prokaryotes
Chapter 28 - Protists
Chapter 29 - Plant Diversity I
Chapter 30 - Plant Diversity II: The Evolution of Seed Plants
Chapter 31 - Fungi
Chapter 32 - An Introduction to Animal Diversity
Chapter 33 - Invertebrates
Chapter 34 - Vertebrates
Chapter 35 - Plant Structure, Growth, and Development
Chapter 36 - Transport in Vascular Plants
Chapter 37 - Plant Nutrition
Chapter 38 - Angiosperm Reproduction and Biotechnology
Chapter 39 - Plant Responses to Internal and External Signals
Chapter 40 - Basic Principles of Animal Form and Function
Chapter 41 - Animal Nutrition
Chapter 42 - Circulation and Gas Exchange
Chapter 43 - The Immune System
Chapter 44 - Osmoregulation and Excretion
Chapter 45 - Hormones and the Endocrine System
Chapter 46 - Animal Reproduction
Chapter 47 - Animal Development
Chapter 48 - Nervous Systems
Chapter 49 - Sensory and Motor Mechanisms
Chapter 50 - An Introduction to Ecology and the Biosphere
Chapter 51 - Behavioral Ecology
Chapter 52 - Population Ecology
Chapter 53 - Community Ecology
Chapter 54 - Ecosystems
Chapter 55 - Conservation Biology and Restoration Ecology
Product details
Language: English ISBN-10: 0805371710
ISBN-13: 9780805371710 978-0805371710
Link download full: Test Bank for Biology 7th edition by Neil Campbell and Jane Reece
https://digitalcontentmarket.org/download/test-bank-for-biology-7th-edition-by-campbell-and-reece/
See more
Test bank for Campbell Biology 10th edition
Test Bank for Microbiology A Human Perspective 7th Edition
You will be guided to the product download page immediately once you complete the payment. If you have any questions, or would like a receive a sample chapter before your purchase, please contact us via email: [email protected]
People also search
Biology 7th edition by Campbell and Reece Test Bank
Test Bank for Biology 7th edition by Campbell and Reece download pdf
Download Test Bank for Biology 7th edition by Campbell and Reece pdf
Download Biology 7th edition by Campbell and Reece Test Bank , guide
Test Bank for Biology 7th edition by Campbell and Reece download free, docs
Instant download Test Bank for Biology 7th edition by Campbell and Reece pdf, answer
Test Bank for Biology 7th edition by Campbell and Reece pdf, ebook
0 notes
Text
Download Test Bank for Biology 7th edition by Campbell and Reece
This is completed downloadable Test Bank for Biology 7th edition by Neil Campbell and Jane Reece
Instant Download Test Bank for Biology 7th edition by Neil Campbell and Jane Reece
View sample:
https://digitalcontentmarket.org/wp-content/uploads/2017/10/Test-Bank-for-Biology-7th-edition-by-Neil-Campbell-and-Jane-Reece.pdf
Product Description:
Neil Campbell and Jane Reece's BIOLOGY remains unsurpassed as the most successful majors biology textbook in the world. This text has invited more than 4 million students into the study of this dynamic and essential discipline.The authors have restructured each chapter around a conceptual framework of five or six big ideas. An Overview draws students in and sets the stage for the rest of the chapter, each numbered Concept Head announces the beginning of a new concept, and Concept Check questions at the end of each chapter encourage students to assess their mastery of a given concept. New Inquiry Figures focus students on the experimental process, and new Research Method Figures illustrate important techniques in biology. Each chapter ends with a Scientific Inquiry Question that asks students to apply scientific investigation skills to the content of the chapter.
Table of Contents:
Chapter 1 - Exploring Life
Chapter 2 - The Chemical Context of Life
Chapter 3 - Water and the Fitness of the Environment
Chapter 4 - Carbon and the Molecular Diversity of Life
Chapter 5 - The Structure and Function of Macromolecules
Chapter 7 - Membrane Structure and Function
Chapter 8 - An Introduction to Metabolism
Chapter 9 - Cellular Respiration: Harvesting Chemical Energy
Chapter 10 - Photosynthesis
Chapter 11- Cell Communication
Chapter 12 - The Cell Cycle
Chapter 13 - Meiosis and Sexual
Chapter 14 - Mendel and the Gene Idea
Chapter 15 - The Chromosomal Basis of Inheritance
Chapter 16 - The Molecular Basis of Inheritance
Chapter 17 - From Gene to Protein
Chapter 18 - The Genetics of Viruses and Bacteria
Chapter 19 - Eukaryotic Genomes: Organization, Regulation, and Evolution
Chapter 20 - DNA Technology
Chapter 21 - The Genetic Basis
Chapter 22 - Descent with Modification: A Darwinian View of Life
Chapter 23 - The Evolution of Populations
Chapter 24 - The Origin of Species
Chapter 25 - Phylogeny and Systematics
Chapter 26 - The Tree of Life
Chapter 27 - Prokaryotes
Chapter 28 - Protists
Chapter 29 - Plant Diversity I
Chapter 30 - Plant Diversity II: The Evolution of Seed Plants
Chapter 31 - Fungi
Chapter 32 - An Introduction to Animal Diversity
Chapter 33 - Invertebrates
Chapter 34 - Vertebrates
Chapter 35 - Plant Structure, Growth, and Development
Chapter 36 - Transport in Vascular Plants
Chapter 37 - Plant Nutrition
Chapter 38 - Angiosperm Reproduction and Biotechnology
Chapter 39 - Plant Responses to Internal and External Signals
Chapter 40 - Basic Principles of Animal Form and Function
Chapter 41 - Animal Nutrition
Chapter 42 - Circulation and Gas Exchange
Chapter 43 - The Immune System
Chapter 44 - Osmoregulation and Excretion
Chapter 45 - Hormones and the Endocrine System
Chapter 46 - Animal Reproduction
Chapter 47 - Animal Development
Chapter 48 - Nervous Systems
Chapter 49 - Sensory and Motor Mechanisms
Chapter 50 - An Introduction to Ecology and the Biosphere
Chapter 51 - Behavioral Ecology
Chapter 52 - Population Ecology
Chapter 53 - Community Ecology
Chapter 54 - Ecosystems
Chapter 55 - Conservation Biology and Restoration Ecology
Product details
Language: English ISBN-10: 0805371710
ISBN-13: 9780805371710 978-0805371710
Link download full: Test Bank for Biology 7th edition by Campbell and Reece
https://digitalcontentmarket.org/download/test-bank-for-biology-7th-edition-by-campbell-and-reece/
See more
Test bank for Campbell Biology 10th edition
Test Bank for Microbiology A Human Perspective 7th Edition
You will be guided to the product download page immediately once you complete the payment. If you have any questions, or would like a receive a sample chapter before your purchase, please contact us via email: [email protected]
People also search
Biology 7th edition by Campbell and Reece Test Bank
Test Bank for Biology 7th edition by Campbell and Reece download pdf
Download Test Bank for Biology 7th edition by Campbell and Reece pdf
Download Biology 7th edition by Campbell and Reece Test Bank , guide
Test Bank for Biology 7th edition by Campbell and Reece download free, docs
Instant download Test Bank for Biology 7th edition by Campbell and Reece pdf, answer
Test Bank for Biology 7th edition by Campbell and Reece pdf, ebook
0 notes
Text
Fwd: Course: Norway_Bergen.SystematicsMarineInvertebrates.Sept26-Oct7
Begin forwarded message: > From: [email protected] > Subject: Course: Norway_Bergen.SystematicsMarineInvertebrates.Sept26-Oct7 > Date: 31 May 2022 at 06:36:23 BST > To: [email protected] > > > ForBio - Research School in Biosystematics and the University of Bergen > Norway (UiB) jointly offer the course: Systematics and Evolution of > Marine Invertebrates > > Time and place: Sep. 26, 2022-Oct. 7, 2022 5:00, Biological Institute > (BIO), University of Bergen > > Course scope: Systematics and Evolution of Marine Invertebrates is a > voyage through the tree-of-life of metazoans offering a comprehensive > overview of the diversity of invertebrate phyla, morphological traits, > and latest hypotheses of evolutionary relationships based on molecular > phylogenetics. A suit of laboratory activities gives students a truly > hands-on experience and opportunity to explore the morphology of a large > ensemble of major representatives of the Animal Kingdom, through the > observation of life and preserved specimens, anatomical dissections, > interpretation of anatomical slides, and optical microscopy. > > Course instructors > > Prof. Manuel Malaquias (University Museum of Bergen, UiB, Norway; > course coordinator) > Ass. Prof. Nataliya Budaeva (University Museum of Bergen, UiB, Norway; > course coordinator) > Prof. Andreas Hejnol (University of Jena, Germany / University of Bergen, > Norway) > Ass. Prof. Aino Hosia (University Museum of Bergen, UiB, Norway) > Dr. Luis Martell (University Museum of Bergen, UiB, Norway) > Prof. Elena Temereva (Lomonosov Moscow State University, Russia) > Dr. Nina Mikkelsen (University Museum of Bergen, UiB, Norway) > Ass. Prof. Andreas Altenburger (The Arсtic University of Norway) > Prof. Henrik Glenner (University of Bergen, Norway) > Dr. Kenneth Meland (University of Bergen, Norway) > Ass. Prof. Nicolas Straube (University Museum of Bergen, UiB, Norway) > Dr. Jon Hestetun (NORCE Norwegian Research Centre AS) > Dr. Antonina Kremenetskaia (P.P. Shirshov Institute of Oceanology, > RAS, Russia) > Dr. Francisca Carvalho (University Museum of Bergen, UiB, Norway) > > Learning outcomes > > 1) To describe the morphology and anatomy of the different phyla of > invertebrate animals and how they are adapted to the living > environment. > 2) Acquire knowledge on taxonomy and phylogeny of marine animals, from > sponges to protochordates. > 3) Explain the concepts and terms that underlie phylogenetic > classifications and hypotheses. > 4) Acquire competences on basic anatomical dissection and drawing > techniques. > 5) Acquire competences to understand and discuss conflictive hypotheses > on the evolution of the Metazoa tree-of-life. > 6) Develop a critical attitude towards scientific literature (papers). > 7) Understand the dynamics of the scholar process that underlies the > "making" of Science. > > Application deadline: June 17th 2022 > > More information and registration: > https://ift.tt/OTV2NYs > > Please feel free to contact [email protected] with any questions. > > > Nataliya Budaeva, Associate Professor > Department of Natural History > Section of Taxonomy and Evolution > University Museum of Bergen > University of Bergen > PB 7800 5020 Bergen > Norway > > https://ift.tt/IlipLjO > > Coordinator of ForBio - Research School in Biosystematics > https://ift.tt/JOtf3hB > > Nataliya Budaeva > via IFTTT
0 notes