Don't wanna be here? Send us removal request.
Text
Assignment_4 Testing a Potential Moderator
Chi-Square Test of Independence
#“libraries”
import pandas import numpy import scipy.stats import seaborn import matplotlib.pyplot as plt
data = pandas.read_csv(‘addhealth_pds.csv’, low_memory=False)
#print ('Converting variables to numeric’)
data['H1SU1’] = pandas.to_numeric(data['H1SU1’], errors='coerce’) data['H1NB6’] = pandas.to_numeric(data['H1NB6’], errors='coerce’)
#print ('Coding missing values’)
data[“H1SU1”] = data[“H1SU1”].replace(6, numpy.nan) data[“H1SU1”] = data[“H1SU1”].replace(9, numpy.nan) data[“H1SU1”] = data[“H1SU1”].replace(8, numpy.nan) data[“H1NB6”] = data[“H1NB6”].replace(6, numpy.nan) data[“H1NB6”] = data[“H1NB6”].replace(8, numpy.nan)
#print ('contingency table of observed counts’)
ct1=pandas.crosstab(data['H1SU1’], data['H1NB6’]) print (ct1)
print ('column percentages’) colsum=ct1.sum(axis=0) colpct=ct1/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs1= scipy.stats.chi2_contingency(ct1) print (cs1)
#print ('set variable types’) data[“H1NB6”] = data[“H1NB6”].astype('category’) data['H1SU1’] = pandas.to_numeric(data['H1SU1’], errors='coerce’)
seaborn.factorplot(x=“H1NB6”, y=“H1SU1”, data=data, kind=“bar”, ci=None) plt.xlabel('Happiness Level Living in Neighbourhood 5=Very Happy’) plt.ylabel('Considered Suicide in Past 12 Months’)
recode1= {1: 1, 2: 2} data['COMP1v2’]= data[“H1NB6”].map(recode1)
#print ('contigency table of observed counts’) ct2=pandas.crosstab(data['H1SU1’], data['COMP1v2’]) print (ct2)
#print ('column percentages’) colsum=ct2.sum(axis=0) colpct=ct2/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs2= scipy.stats.chi2_contingency(ct2) print (cs2)
recode2= {1: 1, 3: 3} data['COMP1v3’]= data[“H1NB6”].map(recode2)
#print ('contigency table of observed counts’) ct3=pandas.crosstab(data['H1SU1’], data['COMP1v3’]) print (ct3)
#print ('column percentages’) colsum=ct3.sum(axis=0) colpct=ct3/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs3= scipy.stats.chi2_contingency(ct3) print (cs3)
recode3= {1: 1, 4: 4} data['COMP1v4’]= data[“H1NB6”].map(recode3)
#print ('contigency table of observed counts’) ct4=pandas.crosstab(data['H1SU1’], data['COMP1v4’]) print (ct4)
print ('column percentages’) colsum=ct4.sum(axis=0) colpct=ct4/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs4= scipy.stats.chi2_contingency(ct4) print (cs4)
recode4= {1: 1, 5: 5} data['COMP1v5’]= data[“H1NB6”].map(recode4)
#print ('contigency table of observed counts’) ct5=pandas.crosstab(data['H1SU1’], data['COMP1v5’]) print (ct5)
#print ('column percentages’) colsum=ct5.sum(axis=0) colpct=ct5/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs5= scipy.stats.chi2_contingency(ct5) print (cs5)
recode5= {2: 2, 3: 3} data['COMP2v3’]= data[“H1NB6”].map(recode5)
#print ('contigency table of observed counts’) ct6=pandas.crosstab(data['H1SU1’], data['COMP2v3’]) print (ct6)
#print ('column percentages’) colsum=ct6.sum(axis=0) colpct=ct6/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs6= scipy.stats.chi2_contingency(ct6) print (cs6)
recode6= {2: 2, 4: 4} data['COMP2v4’]= data[“H1NB6”].map(recode6)
#print ('contigency table of observed counts’) ct7=pandas.crosstab(data['H1SU1’], data['COMP2v4’]) print (ct7)
#print ('column percentages’) colsum=ct7.sum(axis=0) colpct=ct7/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs7= scipy.stats.chi2_contingency(ct7) print (cs7)
recode7= {2: 2, 5: 5} data['COMP2v5’]= data[“H1NB6”].map(recode7)
#print ('contigency table of observed counts’) ct8=pandas.crosstab(data['H1SU1’], data['COMP2v5’]) print (ct8)
#print ('column percentages’) colsum=ct8.sum(axis=0) colpct=ct8/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs8= scipy.stats.chi2_contingency(ct8) print (cs8)
recode8= {3: 3, 4: 4} data['COMP3v4’]= data[“H1NB6”].map(recode8)
#print ('contigency table of observed counts’) ct9=pandas.crosstab(data['H1SU1’], data['COMP3v4’]) print (ct9)
#print ('column percentages’) colsum=ct9.sum(axis=0) colpct=ct9/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs9= scipy.stats.chi2_contingency(ct9) print (cs9)
recode9= {3: 3, 5: 5} data['COMP3v5’]= data[“H1NB6”].map(recode9)
#print ('contigency table of observed counts’) ct10=pandas.crosstab(data['H1SU1’], data['COMP3v5’]) print (ct10)
#print ('column percentages’) colsum=ct10.sum(axis=0) colpct=ct10/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs10= scipy.stats.chi2_contingency(ct10) print (cs10)
recode10= {4: 4, 5: 5} data['COMP4v5’]= data[“H1NB6”].map(recode10)
#print ('contigency table of observed counts’) ct11=pandas.crosstab(data['H1SU1’], data['COMP4v5’]) print (ct11)
#print ('column percentages’) colsum=ct11.sum(axis=0) colpct=ct11/colsum print(colpct)
#print ('chi-square value, p value, expected counts’) cs11= scipy.stats.chi2_contingency(ct11) print (cs11)
OUTPUT
Converting variables to numeric Coding missing values contingency table of observed counts H1NB6 1.0 2.0 3.0 4.0 5.0 H1SU1 0.0 138 287 1142 2016 2023 1.0 55 74 227 284 180 column percentages H1NB6 1.0 2.0 3.0 4.0 5.0 H1SU1 0.0 0.715026 0.795014 0.834186 0.876522 0.918293 1.0 0.284974 0.204986 0.165814 0.123478 0.081707 (122.34711107270866, 1.6837131211401846e-25, 4, array([[ 168.37192655, 314.93401805, 1194.30656707, 2006.50482415, 1921.88266418], [ 24.62807345, 46.06598195, 174.69343293, 293.49517585, 281.11733582]])) /Users/tyler2k/anaconda3/lib/python3.7/site-packages/seaborn/categorical.py:3666: UserWarning: The `factorplot` function has been renamed to `catplot`. The original name will be removed in a future release. Please update your code. Note that the default `kind` in `factorplot` (`'point’`) has changed `'strip’` in `catplot`. warnings.warn(msg) COMP1v2 1.0 2.0 H1SU1 0.0 138 287 1.0 55 74 COMP1v2 1.0 2.0 H1SU1 0.0 0.715026 0.795014 1.0 0.284974 0.204986 (4.0678278591878705, 0.04370744446565526, 1, array([[148.05956679, 276.94043321], [ 44.94043321, 84.05956679]])) COMP1v3 1.0 3.0 H1SU1 0.0 138 1142 1.0 55 227 COMP1v3 1.0 3.0 H1SU1 0.0 0.715026 0.834186 1.0 0.284974 0.165814 (15.43912984309443, 8.52056112101083e-05, 1, array([[ 158.15620999, 1121.84379001], [ 34.84379001, 247.15620999]])) COMP1v4 1.0 4.0 H1SU1 0.0 138 2016 1.0 55 284 column percentages COMP1v4 1.0 4.0 H1SU1 0.0 0.715026 0.876522 1.0 0.284974 0.123478 (38.163564128380244, 6.505592851611984e-10, 1, array([[ 166.755716, 1987.244284], [ 26.244284, 312.755716]])) COMP1v5 1.0 5.0 H1SU1 0.0 138 2023 1.0 55 180 COMP1v5 1.0 5.0 H1SU1 0.0 0.715026 0.918293 1.0 0.284974 0.081707 (80.60217876116656, 2.760549400154315e-19, 1, array([[ 174.07053422, 1986.92946578], [ 18.92946578, 216.07053422]])) COMP2v3 2.0 3.0 H1SU1 0.0 287 1142 1.0 74 227 COMP2v3 2.0 3.0 H1SU1 0.0 0.795014 0.834186 1.0 0.204986 0.165814 (2.783546208781313, 0.09523708259004951, 1, array([[ 298.19017341, 1130.80982659], [ 62.80982659, 238.19017341]])) COMP2v4 2.0 4.0 H1SU1 0.0 287 2016 1.0 74 284 COMP2v4 2.0 4.0 H1SU1 0.0 0.795014 0.876522 1.0 0.204986 0.123478 (17.110228714530386, 3.527182890794104e-05, 1, array([[ 312.43254416, 1990.56745584], [ 48.56745584, 309.43254416]])) COMP2v5 2.0 5.0 H1SU1 0.0 287 2023 1.0 74 180 COMP2v5 2.0 5.0 H1SU1 0.0 0.795014 0.918293 1.0 0.204986 0.081707 (51.44490204835613, 7.363494652526882e-13, 1, array([[ 325.23790952, 1984.76209048], [ 35.76209048, 218.23790952]])) COMP3v4 3.0 4.0 H1SU1 0.0 1142 2016 1.0 227 284 COMP3v4 3.0 4.0 H1SU1 0.0 0.834186 0.876522 1.0 0.165814 0.123478 (12.480541372399653, 0.00041121300778005455, 1, array([[1178.33251567, 1979.66748433], [ 190.66748433, 320.33251567]])) COMP3v5 3.0 5.0 H1SU1 0.0 1142 2023 1.0 227 180 COMP3v5 3.0 5.0 H1SU1 0.0 0.834186 0.918293 1.0 0.165814 0.081707 (58.33047436979929, 2.2158497377240775e-14, 1, array([[1213.01371781, 1951.98628219], [ 155.98628219, 251.01371781]])) COMP4v5 4.0 5.0 H1SU1 0.0 2016 2023 1.0 284 180 COMP4v5 4.0 5.0 H1SU1 0.0 0.876522 0.918293 1.0 0.123478 0.081707 (20.793289909858167, 5.116190394045173e-06, 1, array([[2063.00244282, 1975.99755718], [ 236.99755718, 227.00244282]]))
Generating a Correlation Coefficient
Code
import pandas import numpy import seaborn import scipy import matplotlib.pyplot as plt
data = pandas.read_csv(‘addhealth_pds.csv’, low_memory=False)
“converting variables to numeric” data[“H1SU2”] = data[“H1SU2”].convert_objects(convert_numeric=True) data[“H1WP8”] = data[“H1WP8”].convert_objects(convert_numeric=True)
“Coding missing values”
data[“H1SU2”] = data[“H1SU2”].replace(6, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(7, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(8, numpy.nan)
data[“H1WP8”] = data[“H1WP8”].replace(96, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(97, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(98, numpy.nan)
scat1 = seaborn.regplot(x='H1SU2’, y='H1WP8’, fit_reg=True, data=data) plt.xlabel('Number of suicide attempts in past 12 months’) plt.ylabel('how many of the past 7 days there was at least one parent in the room with the respondent for their evening meal’) plt.title('Scatterplot for the association between respondents suicide attemps and how many days they ate their evening meal with their parents’)
data_clean=data.dropna()
print ('association between H1SU2 and H1WP8’) print (scipy.stats.pearsonr(data_clean['H1SU2’], data_clean['H1WP8’]))
ANOVA
#post hoc ANOVA
import pandas
import numpy
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
data = pandas.read_csv(‘addhealth_pds.csv’, low_memory=False)
print(“converting variables to numeric”)
data[“H1SU1”] = data[“H1SU1”].convert_objects(convert_numeric=True)
data[“H1NB5”] = data[“H1NB5”].convert_objects(convert_numeric=True)
data[“H1NB6”] = data[“H1NB6”].convert_objects(convert_numeric=True)
print(“Coding missing values”)
data[“H1SU1”] = data[“H1SU1”].replace(6, numpy.nan)
data[“H1SU1”] = data[“H1SU1”].replace(9, numpy.nan)
data[“H1SU1”] = data[“H1SU1”].replace(8, numpy.nan)
data[“H1NB5”] = data[“H1NB5”].replace(6, numpy.nan)
data[“H1NB6”] = data[“H1NB6”].replace(6, numpy.nan)
data[“H1NB6”] = data[“H1NB6”].replace(8, numpy.nan)
#F-Statistic
model1 = smf.ols(formula=‘H1SU1 ~ C(H1NB6)’, data=data)
results1 = model1.fit()
print (results1.summary())
sub1 = data[['H1SU1’, 'H1NB6’]].dropna()
print ('means for H1SU1 by happiness level in neighbourhood’)
m1= sub1.groupby('H1NB6’).mean()
print (m1)
print ('standard deviation for H1SU1 by happiness level in neighbourhood’)
sd1 = sub1.groupby('H1NB6’).std()
print (sd1)
#more tahn 2 lvls
sub2 = sub1[['H1SU1’, 'H1NB6’]].dropna()
model2 = smf.ols(formula='H1SU1 ~ C(H1NB6)’, data=sub2).fit()
print (model2.summary())
print ('2: means for H1SU1 by happiness level in neighbourhood’)
m2= sub2.groupby('H1NB6’).mean()
print (m2)
print ('2: standard deviation for H1SU1 by happiness level in neighbourhood’)
sd2 = sub2.groupby('H1NB6’).std()
print (sd2)
mc1 = multi.MultiComparison(sub2['H1SU1’], sub2 ['H1NB6’])
res1 = mc1.tukeyhsd()
print(res1.summary())
ANOVA RESULTS
converting variables to numeric
Coding missing values
OLS Regression Results
==============================================================================
Dep. Variable: H1SU1 R-squared: 0.019
Model: OLS Adj. R-squared: 0.018
Method: Least Squares F-statistic: 31.16
Date: Sat, 24 Aug 2019 Prob (F-statistic): 9.79e-26
Time: 17:16:11 Log-Likelihood: -2002.8
No. Observations: 6426 AIC: 4016.
Df Residuals: 6421 BIC: 4049.
Df Model: 4
Covariance Type: nonrobust
===================================================================================
coef std err t P>|t| [0.025 0.975]
———————————————————————————–
Intercept 0.2850 0.024 11.976 0.000 0.238 0.332
C(H1NB6)[T.2.0] -0.0800 0.029 -2.713 0.007 -0.138 -0.022
C(H1NB6)[T.3.0] -0.1192 0.025 -4.688 0.000 -0.169 -0.069
C(H1NB6)[T.4.0] -0.1615 0.025 -6.519 0.000 -0.210 -0.113
C(H1NB6)[T.5.0] -0.2033 0.025 -8.191 0.000 -0.252 -0.155
==============================================================================
Omnibus: 2528.245 Durbin-Watson: 1.952
Prob(Omnibus): 0.000 Jarque-Bera (JB): 7313.190
Skew: 2.173 Prob(JB): 0.00
Kurtosis: 5.903 Cond. No. 15.0
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
means for H1SU1 by happiness level in neighbourhood
H1SU1
H1NB6
1.0 0.284974
2.0 0.204986
3.0 0.165814
4.0 0.123478
5.0 0.081707
standard deviation for H1SU1 by happiness level in neighbourhood
H1SU1
H1NB6
1.0 0.452576
2.0 0.404252
3.0 0.372050
4.0 0.329057
5.0 0.273980
/For all other conversions use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.
data[“H1SU1”] = data[“H1SU1”].convert_objects(convert_numeric=True)
For all other conversions use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.
data[“H1NB5”] = data[“H1NB5”].convert_objects(convert_numeric=True)
For all other conversions use the data-type specific converters pd.to_datetime, pd.to_timedelta and pd.to_numeric.
data[“H1NB6”] = data[“H1NB6”].convert_objects(convert_numeric=True)
OLS Regression Results
==============================================================================
Dep. Variable: H1SU1 R-squared: 0.019
Model: OLS Adj. R-squared: 0.018
Method: Least Squares F-statistic: 31.16
Date: Sat, 24 Aug 2019 Prob (F-statistic): 9.79e-26
Time: 17:16:11 Log-Likelihood: -2002.8
No. Observations: 6426 AIC: 4016.
Df Residuals: 6421 BIC: 4049.
Df Model: 4
Covariance Type: nonrobust
===================================================================================
coef std err t P>|t| [0.025 0.975]
———————————————————————————–
Intercept 0.2850 0.024 11.976 0.000 0.238 0.332
C(H1NB6)[T.2.0] -0.0800 0.029 -2.713 0.007 -0.138 -0.022
C(H1NB6)[T.3.0] -0.1192 0.025 -4.688 0.000 -0.169 -0.069
C(H1NB6)[T.4.0] -0.1615 0.025 -6.519 0.000 -0.210 -0.113
C(H1NB6)[T.5.0] -0.2033 0.025 -8.191 0.000 -0.252 -0.155
==============================================================================
Omnibus: 2528.245 Durbin-Watson: 1.952
Prob(Omnibus): 0.000 Jarque-Bera (JB): 7313.190
Skew: 2.173 Prob(JB): 0.00
Kurtosis: 5.903 Cond. No. 15.0
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
2: means for H1SU1 by happiness level in neighbourhood
H1SU1
H1NB6
1.0 0.284974
2.0 0.204986
3.0 0.165814
4.0 0.123478
5.0 0.081707
2: standard deviation for H1SU1 by considered suicide in past 12 months
H1SU1
H1NB6
1.0 0.452576
2.0 0.404252
3.0 0.372050
4.0 0.329057
5.0 0.273980
Multiple Comparison of Means - Tukey HSD,FWER=0.05
=============================================
group1 group2 meandiff lower upper reject
———————————————
1.0 2.0 -0.08 -0.1604 0.0004 False
1.0 3.0 -0.1192 -0.1885 -0.0498 True
1.0 4.0 -0.1615 -0.2291 -0.0939 True
1.0 5.0 -0.2033 -0.271 -0.1356 True
2.0 3.0 -0.0392 -0.0925 0.0142 False
2.0 4.0 -0.0815 -0.1326 -0.0304 True
2.0 5.0 -0.1233 -0.1745 -0.0721 True
3.0 4.0 -0.0423 -0.0731 -0.0115 True
3.0 5.0 -0.0841 -0.1152 -0.0531 True
4.0 5.0 -0.0418 -0.0687 -0.0149 True
0 notes
Text
Assignment_3 Income with consumption of alcohol correlation coefficient

incomeperperson and alcconsumption, the correlation coefficient (r) is approximately 0.30 with a p-value of 0.0001.
relationship is statistically significant.
weak and positive
0 notes
Text
Assignment 1
import pandas import numpy import seaborn import scipy import matplotlib.pyplot as plt
data = pandas.read_csv(‘addhealth_pds.csv’, low_memory=False)
“converting variables to numeric” data[“H1SU2”] = data[“H1SU2”].convert_objects(convert_numeric=True) data[“H1WP8”] = data[“H1WP8”].convert_objects(convert_numeric=True)
“Coding missing values”
data[“H1SU2”] = data[“H1SU2”].replace(6, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(7, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(8, numpy.nan)
data[“H1WP8”] = data[“H1WP8”].replace(96, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(97, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(98, numpy.nan)
scat1 = seaborn.regplot(x=‘H1SU2’, y='H1WP8’, fit_reg=True, data=data) plt.xlabel('Number of suicide attempts in past 12 months’) plt.ylabel('how many of the past 7 days there was at least one parent in the room with the respondent for their evening meal’) plt.title('Scatterplot for the association between respondents suicide attemps and how many days they ate their evening meal with their parents’)
data_clean=data.dropna()
print ('association between H1SU2 and H1WP8’) print (scipy.stats.pearsonr(data_clean['H1SU2’], data_clean['H1WP8’]))
Results association between H1SU2 and H1WP8 (-0.040373727372780416, 0.25432562116292673)
Analysis Using the Pearson correlation an association between the number of respondents’ suicide attempts within the last 12 months were measured. Also within the last 7 they ate their evening meal with one of their parents being present for how many days . The more a respondent ate with their parents, the closer their bond is, and therefore the less they would attempt suicide since they have a positive relationship with their family.
The association was very low as I only got a P-value of -0.04 or a 4% negative correlation between both values. As such, it is concluded that the number of meals that respondents’ eat with their parents does not have a large impact on the amount of suicide attempts that they might have.
1 note
·
View note
Text
Assignment 1
import pandas import numpy import seaborn import scipy import matplotlib.pyplot as plt data = pandas.read_csv(‘addhealth_pds.csv’, low_memory=False) “converting variables to numeric” data[“H1SU2”] = data[“H1SU2”].convert_objects(convert_numeric=True) data[“H1WP8”] = data[“H1WP8”].convert_objects(convert_numeric=True) “Coding missing values” data[“H1SU2”] = data[“H1SU2”].replace(6, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(7, numpy.nan) data[“H1SU2”] = data[“H1SU2”].replace(8, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(96, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(97, numpy.nan) data[“H1WP8”] = data[“H1WP8”].replace(98, numpy.nan) scat1 = seaborn.regplot(x='H1SU2’, y='H1WP8’, fit_reg=True, data=data) plt.xlabel('Number of suicide attempts in past 12 months’) plt.ylabel('how many of the past 7 days there was at least one parent in the room with the respondent for their evening meal’) plt.title('Scatterplot for the association between respondents suicide attemps and how many days they ate their evening meal with their parents’) data_clean=data.dropna() print ('association between H1SU2 and H1WP8’) print (scipy.stats.pearsonr(data_clean['H1SU2’], data_clean['H1WP8’])) Results association between H1SU2 and H1WP8 (-0.040373727372780416, 0.25432562116292673) Analysis Using the Pearson correlation an association between the number of respondents’ suicide attempts within the last 12 months were measured. Also within the last 7 they ate their evening meal with one of their parents being present for how many days . The more a respondent ate with their parents, the closer their bond is, and therefore the less they would attempt suicide since they have a positive relationship with their family. The association was very low as I only got a P-value of -0.04 or a 4% negative correlation between both values. As such, it is concluded that the number of meals that respondents’ eat with their parents does not have a large impact on the amount of suicide attempts that they might have.
1 note
·
View note
Text
Chi Square test
import pandas import numpy import scipy.stats import seaborn import matplotlib.pyplot as plt
# read data ‘nesarc in python
data1=pandas.read_csv('my_nesarc.csv’, low_memory=False)
#worked" convert to numeric as python read data as object(string)
data1['S2DQ1’]=pandas.to_numeric(data1['S2DQ1’], errors='coerce’)
data1['S2AQ3’]=pandas.to_numeric(data1['S2AQ3’], errors='coerce’)
data1['MARITAL’]=pandas.to_numeric(data1['MARITAL’], errors='coerce’)
data1.dtypes Out[8]: Unnamed: 0 int64 ETHRACE2A int64 ETOTLCA2 object IDNUM int64 PSU int64
HER12ABDEP int64 HERP12ABDEP int64 OTHB12ABDEP int64 OTHBP12ABDEP int64 NDSymptoms float64 Length: 3010, dtype: object
#replacing missing values for nan (ALSO 9 OR 99 CONSIDERING MISSING)
print('count for original S2DQ1’) count for original S2DQ1
f1=data1['S2DQ1’].value_counts(sort=False,dropna=False)
print(f1) 1 8124 2 32445 9 2524 Name: S2DQ1, dtype: int64
print('count for S2DQ1 by replacing missing with nan’) count for S2DQ1 by replacing missing with nan
data1['S2DQ1’]=data1['S2DQ1’].replace(9, numpy.nan)
f11=data1['S2DQ1’].value_counts(sort=False, dropna=False)
print(f11) 1.0 8124 2.0 32445 NaN 2524 Name: S2DQ1, dtype: int64
print('count for original S2AQ3’) count for original S2AQ3
f2=data1['S2AQ3’].value_counts(sort=False,dropna=False)
print(f2) 1 26946 2 16116 9 31 Name: S2AQ3, dtype: int64
print('count for S2AQ3 by replacing missing with nan’) count for S2AQ3 by replacing missing with nan
data1['S2AQ3’]=data1['S2AQ3’].replace(9,numpy.nan)
f22=data1['S2AQ3’].value_counts(sort=False, dropna=False)
print(f22) 2.0 16116 1.0 26946 NaN 31 Name: S2AQ3, dtype: int64
print('count for original MARITAL’) count for original MARITAL
f3=data1['MARITAL’].value_counts(sort=False,dropna=False)
print(f3) # no missing values 1 20769 2 1312 3 4271 4 5401 5 1445 6 9895 Name: MARITAL, dtype: int64
# select rows: subset of data1 given that FATHER EVER AN ALCOHOLIC OR PROBLEM DRINKER’
sub1=data1[(data1['S2DQ1’]==1)]
sub2=sub1[['MARITAL’,'S2AQ3’]] 8124
sub3=sub2.dropna()
print(len(sub3)) 8123
#frequency
f4=sub3['S2AQ3’].value_counts()
print(f4) 1.0 5544 2.0 2579 Name: S2AQ3, dtype: int64
f5=sub3['MARITAL’].value_counts()
print(f5) 1 3837 6 1871 4 1263 3 470 2 351 5 331 Name: MARITAL, dtype: int64
# contingency table of observed counts
cto=pandas.crosstab(sub3['S2AQ3’], sub3['MARITAL’], margins=True)
print (cto) MARITAL 1 2 3 4 5 6 All S2AQ3 1.0 2571 274 206 869 216 1408 5544 2.0 1266 77 264 394 115 463 2579 All 3837 351 470 1263 331 1871 8123
# column percentages
colsum=cto.sum(axis=0)
colpt=cto/colsum
MARITAL 1 2 3 4 5 6 All S2AQ3 1.0 0.335027 0.390313 0.219149 0.344022 0.326284 0.376269 0.341253 2.0 0.164973 0.109687 0.280851 0.155978 0.173716 0.123731 0.158747 All 0.500000 0.500000 0.500000 0.500000 0.500000 0.500000 0.500000
print(colpt)
# chi-square
print ('chi-square value, p value, expected counts’) chi-square value, p value, expected counts
cs1= scipy.stats.chi2_contingency(cto)
print (cs1) (191.58929962177555, 1.7679125907777407e-34, 12, array([[2618.77729903, 239.55976856, 320.77803767, 862.00566293, 225.9096393 , 1276.96959252, 5544. ], [1218.22270097, 111.44023144, 149.22196233, 400.99433707, 105.0903607 , 594.03040748, 2579. ], [3837. , 351. , 470. , 1263. , 331. , 1871. , 8123. ]]))
Conclusion: as the p value is very small; drinking alcohol depend on marital status with family history of drinking For chi square post hoc paired comparisons test
0 notes
Photo

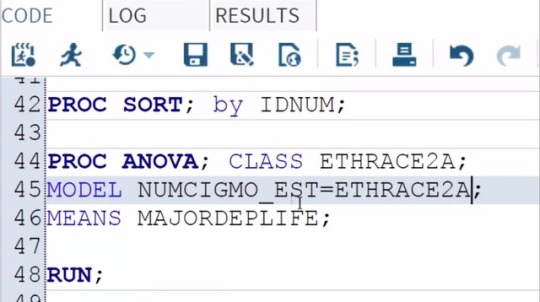
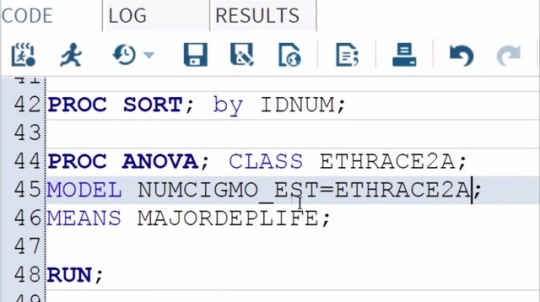
Data Analysis Tool: Assignment_1
In order to conduct post hoc paired comparisons in the context of Mironova.
Examining the association between ethnicity and number of cigarettes smoked
per month among young adult smokers, I'm going to use the Duncan Test.
To do this, all I need to do is add a slash and
the word Duncan at the end of my MEANS statement.
And then save and run my program.
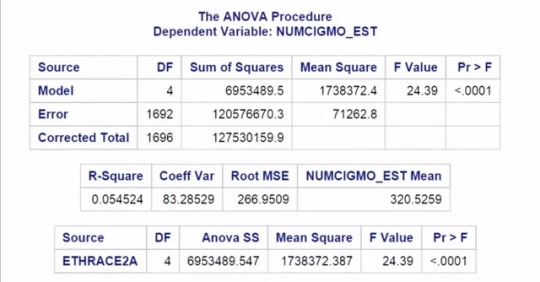
Here are the Proc ANOVA results with Duncan Post Hoc Test.
The top of our results looks the same as in our original test.
The F value or F statistic is 24.4 and
it's significant at the P < 0.0001 level.
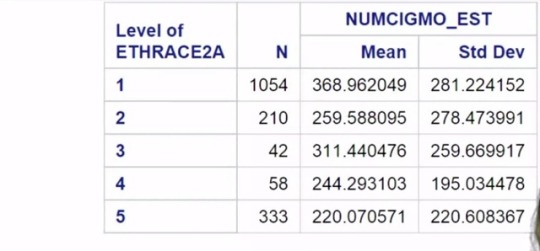
However, if we scroll down, we see a new table displaying the results of the paired
comparisons conducted by the Duncan Multiple Range Test.
1 note
·
View note