Don't wanna be here? Send us removal request.
Text
Data Management and Visualization Assignment 4
describe of patients in junior_young_patients group count 9 unique 2 top male freq 6 Name: gender, dtype: object

describe of patients in senior_young_patients group count 8 unique 2 top male freq 4

describe of patients in junior_adults_patients group count 25 unique 2 top female freq 13

describe of patients in senior_adults_patients group count 32 unique 2 top male freq 17

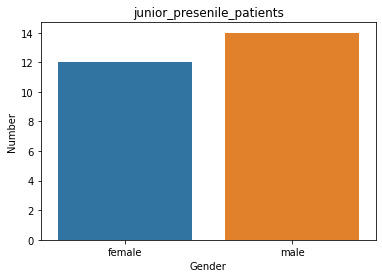
describe of patients in junior_presenile_patients group count 26 unique 2 top male freq 14
describe of patients in senior_presenile_patients group count 28 unique 2 top male freq 16

describe of patients in junior_elderly_patients group count 29 unique 2 top male freq 15

describe of patients in senior_elderly_patients group count 17 unique 2 top male freq 9

describe of patients in old_patients group
count 11 unique 2 top male freq 6

0 notes
Text
Data Management and Visualization - Week 2/3 Assignment
Summary:
I filtered the data and cleaned it from NaN, the following code display three of many variables that I will use.
I divided the patients to groups by age: junior_young_paients ages = 1-19 senior_young_paients ages = 20-30 junior_adults_paients ages = 31-40 senior_adults_paients ages = 41-55 junior_presenile_paients ages = 56-67 senior_presenile_paients ages = 68-75 junior_elderly_paients ages = 76-85 senior_elderly_paients ages = 86-110
The 3 variables are:
1) Number of patients cataloged by males and females inside each group
2) Number of patients cataloged by country inside each group
3) Number of deaths cataloged by country inside each group
Code:
# -*- coding: utf-8 -*- """ Created on Fri Mar 27 12:02:46 2020
@author: RaziSh """
import pandas as pd import numpy as np
########### Getting data from dataset ############
data = pd.read_csv('G:/Courses/Data_Management_and_Visualization/DataSet/COVID19_line_list_data.csv', low_memory=False) #print (len(data)) #number of observations (rows) #print (len(data.columns)) # number of variables (columns)
########### Dropping NaN ############
#setting variables you will be working with to numeric data = data[data['age'].notna()] #print (len(data)) #number of observations (rows) #print (len(data.columns)) # number of variables (columns)
########### Extracting the needed data from dataset ############
data['age'] = pd.to_numeric(data['age']) data['visiting Wuhan'] = pd.to_numeric(data['visiting Wuhan']) data['from Wuhan'] = pd.to_numeric(data['from Wuhan']) data.loc[data['death'] != ("1" or "0" ), 'death'] = 0 data['death'] = pd.to_numeric(data['death']) data.loc[data['recovered'] != ("1" or "0" ), 'recovered'] = 0 data['recovered'] = pd.to_numeric(data['recovered'])
#print ("Full ages distribution") #age_distribution= data.groupby('age').size() #print (age_distribution) #print ("Full ages distribution by %") #perc_age_distribution = data.groupby('age').size() * 100 / len(data) #print (perc_age_distribution)
########### Defining groups ############ print("The patients are divided to groups by ages:") print("junior_young_paients ages = 1-19 ") print("senior_young_paients ages = 20-30 ") print("junior_adults_paients ages = 31-40 ") print("senior_adults_paients ages = 41-55 ") print("junior_presenile_paients ages = 56-67 ") print("senior_presenile_paients ages = 68-75 ") print("junior_elderly_paients ages = 76-85 ") print("senior_elderly_paients ages = 86-110 ")
########### Making subdata from the extracted data ############
#subset data to young age 1 to 19 who have illed and dead grouped by gender junior_young_paients=data[(data['age']>=1) & (data['age']<=19)].groupby(['country','gender']).size().reset_index(name='Patients No.') junior_young_paients_death=data[(data['age']>=1) & (data['age']<=19) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 20 to 30 who have illed and dead grouped by gender senior_young_paients=data[(data['age']>=20) & (data['age']<=30)].groupby(['country','gender']).size().reset_index(name='Patients No.') senior_young_paients_death=data[(data['age']>=20) & (data['age']<=30) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 31 to 40 who have illed and dead grouped by gender junior_adults_paients=data[(data['age']>=31) & (data['age']<=40)].groupby(['country','gender']).size().reset_index(name='Patients No.') junior_adults_paients_deaths=data[(data['age']>=31) & (data['age']<=40) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 41 to 55 who have illed and dead grouped by gender senior_adults_paients=data[(data['age']>=41) & (data['age']<=55)].groupby(['country','gender']).size().reset_index(name='Patients No.') senior_adults_paients_death=data[(data['age']>=41) & (data['age']<=55) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 56 to 67 who have illed and dead grouped by gender junior_presenile_paients=data[(data['age']>=56) & (data['age']<=67)].groupby(['country','gender']).size().reset_index(name='Patients No.') junior_presenile_paients_deaths=data[(data['age']>=56) & (data['age']<=67) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 68 to 75 who have illed and dead grouped by gender senior_presenile_paients=data[(data['age']>=68) & (data['age']<=75)].groupby(['country','gender']).size().reset_index(name='Patients No.') senior_presenile_paients_death=data[(data['age']>=68) & (data['age']<=75) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 76 to 85 who have illed and dead grouped by gender junior_elderly_paients=data[(data['age']>=76) & (data['age']<=85)].groupby(['country','gender']).size().reset_index(name='Patients No.') junior_elderly_paients_death=data[(data['age']>=76) & (data['age']<=85) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
#subset data to young age 86 to 110 who have illed and dead grouped by gender senior_elderly_paients=data[(data['age']>=86) & (data['age']<=110)].groupby(['country','gender']).size().reset_index(name='Patients No.') senior_elderly_paients_death=data[(data['age']>=86) & (data['age']<=110) & (data['death']== 1)].groupby(['country']).size().reset_index(name='deaths')
########### Distbrution of paients inside the groups by gender ############
print ('Number of males/females inside the junior_young_paients:') internal_junior_young_paients_distribution = junior_young_paients.groupby('gender').sum() internal_junior_young_paients_distribution = internal_junior_young_paients_distribution.reset_index() print(internal_junior_young_paients_distribution)
print ('Number of males/females inside the senior_young_paients:') internal_senior_young_paients_distribution = senior_young_paients.groupby('gender').sum() internal_senior_young_paients_distribution = internal_senior_young_paients_distribution.reset_index() print(internal_senior_young_paients_distribution)
print ('Number of males/females inside the junior_adults_paients:') internal_junior_adults_paients_distribution = junior_adults_paients.groupby('gender').sum() internal_junior_adults_paients_distribution = internal_junior_adults_paients_distribution.reset_index() print(internal_junior_young_paients_distribution)
print ('counts for males/females inside the senior_adults_paients:') internal_senior_adults_paients_distribution = senior_adults_paients.groupby('gender').sum() internal_senior_adults_paients_distribution = internal_senior_adults_paients_distribution.reset_index() print(internal_senior_adults_paients_distribution)
print ('Number of males/females inside the junior_presenile_paients:') internal_junior_presenile_paients_distribution = junior_presenile_paients.groupby('gender').sum() internal_junior_presenile_paients_distribution = internal_junior_presenile_paients_distribution.reset_index() print(internal_junior_presenile_paients_distribution)
print ('Number of males/females inside the senior_presenile_paients:') internal_senior_presenile_paients_distribution = senior_presenile_paients.groupby('gender').sum() internal_senior_presenile_paients_distribution = internal_senior_presenile_paients_distribution.reset_index() print(internal_senior_presenile_paients_distribution)
print ('Number of males/females inside the junior_elderly_paients:') internal_junior_elderly_paients_distribution = junior_elderly_paients.groupby('gender').sum() internal_junior_elderly_paients_distribution = internal_junior_elderly_paients_distribution.reset_index() print(internal_junior_elderly_paients_distribution)
print ('Number of males/females inside the senior_elderly_paients:') internal_senior_elderly_paients_distribution = senior_elderly_paients.groupby('gender').sum() internal_senior_elderly_paients_distribution = internal_senior_elderly_paients_distribution.reset_index() print(internal_senior_elderly_paients_distribution)
########### Distbrution of paients inside the groups by country ############
print ('Number of paients inside the junior_young_paients by country:') internal_junior_young_paients_distribution = junior_young_paients.groupby('country').sum() internal_junior_young_paients_distribution = internal_junior_young_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_junior_young_paients_distribution)
print ('Number of paients inside the senior_young_paients by country:') internal_senior_young_paients_distribution = senior_young_paients.groupby('country').sum() internal_senior_young_paients_distribution = internal_senior_young_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_senior_young_paients_distribution)
print ('Number of paients inside the junior_adults_paients by country:') internal_junior_adults_paients_distribution = junior_adults_paients.groupby('country').sum() internal_junior_adults_paients_distribution = internal_junior_adults_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_junior_young_paients_distribution)
print ('Number of paients inside the senior_adults_paients by country:') internal_senior_adults_paients_distribution = senior_adults_paients.groupby('country').sum() internal_senior_adults_paients_distribution = internal_senior_adults_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_senior_adults_paients_distribution)
print ('Number of paients inside the junior_presenile_paients by country:') internal_junior_presenile_paients_distribution = junior_presenile_paients.groupby('country').sum() internal_junior_presenile_paients_distribution = internal_junior_presenile_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_junior_presenile_paients_distribution)
print ('Number of paients inside the senior_presenile_paients by country:') internal_senior_presenile_paients_distribution = senior_presenile_paients.groupby('country').sum() internal_senior_presenile_paients_distribution = internal_senior_presenile_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_senior_presenile_paients_distribution)
print ('Number of paients inside the junior_elderly_paients by country:') internal_junior_elderly_paients_distribution = junior_elderly_paients.groupby('country').sum() internal_junior_elderly_paients_distribution = internal_junior_elderly_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_junior_elderly_paients_distribution)
print ('Number of paients inside the senior_elderly_paients by country:') internal_senior_elderly_paients_distribution = senior_elderly_paients.groupby('country').sum() internal_senior_elderly_paients_distribution = internal_senior_elderly_paients_distribution.sort_values('Patients No.', ascending=False) print(internal_senior_elderly_paients_distribution)
########### Distbrution of death inside the groups by country ############
print ('Number of deaths inside the junior_young_paients by country:') if(junior_young_paients_death.empty): print("No deaths in this group.") else: print(junior_young_paients_death)
print ('Number of deaths inside the senior_young_paients by country:') if(senior_young_paients_death.empty): print("No deaths in this group.") else: print(senior_young_paients_death)
print ('Number of deaths inside the junior_adults_paients by country:') if(junior_adults_paients_deaths.empty): print("No deaths in this group.") else: print(junior_adults_paients_deaths)
print ('Number of deaths inside the senior_adults_paients by country:') if(senior_adults_paients_death.empty): print("No deaths in this group.") else: print(senior_adults_paients_death)
print ('Number of deaths inside the junior_presenile_paients by country:') if(junior_presenile_paients_deaths.empty): print("No deaths in this group.") else: print(junior_presenile_paients_deaths)
print ('Number of deaths inside the senior_presenile_paients by country:') if(senior_presenile_paients_death.empty): print("No deaths in this group.") else: print(senior_presenile_paients_death)
print ('Number of deaths inside the junior_elderly_paients by country:') if(junior_elderly_paients_death.empty): print("No deaths in this group.") else: print(junior_elderly_paients_death)
print ('Number of deaths inside the senior_elderly_paients by country:') if(senior_elderly_paients_death.empty): print("No deaths in this group.") else: print(senior_elderly_paients_death)
Output:
The patients are divided to groups by ages: junior_young_paients ages = 1-19 senior_young_paients ages = 20-30 junior_adults_paients ages = 31-40 senior_adults_paients ages = 41-55 junior_presenile_paients ages = 56-67 senior_presenile_paients ages = 68-75 junior_elderly_paients ages = 76-85 senior_elderly_paients ages = 86-110 Number of males/females inside the junior_young_paients: gender Patients No. 0 female 11 1 male 15 Number of males/females inside the senior_young_paients: gender Patients No. 0 female 49 1 male 55 Number of males/females inside the junior_adults_paients: gender Patients No. 0 female 11 1 male 15 counts for males/females inside the senior_adults_paients: gender Patients No. 0 female 88 1 male 147 Number of males/females inside the junior_presenile_paients: gender Patients No. 0 female 81 1 male 94 Number of males/females inside the senior_presenile_paients: gender Patients No. 0 female 40 1 male 50 Number of males/females inside the junior_elderly_paients: gender Patients No. 0 female 15 1 male 25 Number of males/females inside the senior_elderly_paients: gender Patients No. 0 female 4 1 male 4 Number of paients inside the junior_young_paients by country: Patients No. country China 9 Japan 5 Malaysia 4 Singapore 2 Australia 1 Hong Kong 1 South Korea 1 Thailand 1 UAE 1 Vietnam 1 Number of paients inside the senior_young_paients by country: Patients No. country China 21 South Korea 20 Japan 15 Singapore 12 Hong Kong 10 Spain 6 Taiwan 4 Vietnam 3 Australia 2 Canada 2 France 2 Germany 2 Thailand 2 Malaysia 1 Sweden 1 USA 1 Number of paients inside the junior_adults_paients by country: Patients No. country China 9 Japan 5 Malaysia 4 Singapore 2 Australia 1 Hong Kong 1 South Korea 1 Thailand 1 UAE 1 Vietnam 1 Number of paients inside the senior_adults_paients by country: Patients No. country Japan 70 China 49 Singapore 28 South Korea 20 Taiwan 16 Hong Kong 16 Australia 6 Germany 5 Malaysia 5 Spain 4 Canada 4 Vietnam 3 France 3 Thailand 2 Lebanon 1 UAE 1 UK 1 Phillipines 1 Number of paients inside the junior_presenile_paients by country: Patients No. country Japan 39 China 35 Hong Kong 28 South Korea 26 Singapore 15 Malaysia 5 France 4 USA 4 Australia 3 Taiwan 3 Thailand 3 Canada 2 Germany 2 Spain 2 UAE 1 Phillipines 1 Cambodia 1 Vietnam 1 Number of paients inside the senior_presenile_paients by country: Patients No. country Japan 29 China 18 Hong Kong 18 South Korea 8 Thailand 5 Singapore 4 Taiwan 3 UAE 2 France 1 Spain 1 Switzerland 1 Number of paients inside the junior_elderly_paients by country: Patients No. country China 13 Japan 13 Hong Kong 6 South Korea 3 Taiwan 2 France 1 Malaysia 1 Singapore 1 Number of paients inside the senior_elderly_paients by country: Patients No. country China 4 Hong Kong 4 Number of deaths inside the junior_young_paients by country: No deaths in this group. Number of deaths inside the senior_young_paients by country: No deaths in this group. Number of deaths inside the junior_adults_paients by country: country deaths 0 China 1 1 Hong Kong 1 Number of deaths inside the senior_adults_paients by country: country deaths 0 China 3 Number of deaths inside the junior_presenile_paients by country: country deaths 0 China 12 1 Taiwan 1 Number of deaths inside the senior_presenile_paients by country: country deaths 0 China 8 1 Hong Kong 1 Number of deaths inside the junior_elderly_paients by country: country deaths 0 China 11 Number of deaths inside the senior_elderly_paients by country: country deaths 0 China 4
0 notes
Text
Corona virus
I decide that I am most interested in exploring the association between Corona virus spreading and air transport dependence.
Hypothesis:
The more the city/country/continent aviation is developed and have more passengers, the spreading will be quicker and more lethal.
Lower death rate estimates for Corona virus, especially for non-elderly.
Codebook(variables considered):
Sno - Serial number
Province/State - Province or state of the observation.
Country/Region - Country of observation
Confirmed - Cumulative number of confirmed cases till that date
Deaths - Cumulative number of deaths till that date
Recovered - Cumulative number of recovered cases till that date
TopAirports – ranked by aircraft departures, passengers and volume of freight.
International Route Area – Traveling from X to Y continent
Airport Pair – Traveling from X to Y airport
PassengersNo – Cumulative number of passengers in 2018 between two airports
AvgAge – Average age of the people died.
References:
https://www.iata.org/contentassets/a686ff624550453e8bf0c9b3f7f0ab26/wats-2019-mediakit.pdf
https://www.icao.int/sustainability/Documents/MonthlyMonitor-2020/MonthlyMonitor_January2020.pdf
https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset/tasks?taskId=508
0 notes