Don't wanna be here? Send us removal request.
Text
Running a k-means Cluster Analysis(HW4)
In this homework I have conducted k-means cluster analysis to perform grouping of fetals based on some similarities in some characteristics, that could impact their health. These are:
Baseline value
Uterine_contractions
Light_decelerations
Severe_decelerations
Prolongued_decelerations
Abnormal_short_term_variability
Mean_value_of_short_term_variability
Histogram_mode
Histogram_mean
Histogram_median
Histogram_variance
All clustering variables were standardized to have a mean of 0 and a standard deviation of 1 in order to balance all scales.
Then I have randomly split data into train and test splits (70/30) to train and test my k-means model. In order to test influence of cluster number and select the best number of clusters, I have conducted series of analysis, fitting model with k=1-9 clusters. The variance in the clustering variables that was accounted for by the clusters (r-square) was plotted for each of the nine cluster solutions in an elbow curve to provide guidance for choosing the number of clusters to interpret. Results can be observed below:

Results for k=2,4 and 7can be interpreted due to the presence of a fracture point in this positions. I’ve selected k=4 for my further analysis
To reduce number of variables PCA analysis were performed.
A scatterplot of the first two canonical variables by cluster (Figure 2 shown below) can be seen below:
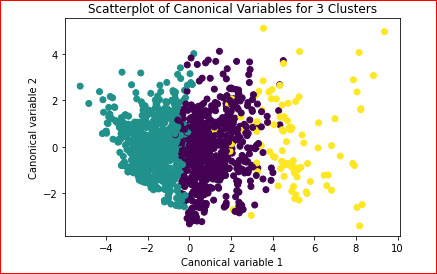
Cluster with green dots has low cluster variance somewhat, cluster with purple dots is also packed well enough, with some variance exists.
cluster with yellow dots isn't well separated from purple and Somewhat from cluster with purple , but it is much more spread on the plot
cluster with yellow it is much more spread on the plot, showing high variance in the plot. data is well separated (clusters overlap is not significant) k=>4 is a suitable number for current situation.
Cluster 0, had the highest Likelihood to be affected by Baseline value.
Cluster 3, includes fetals with higher likelihood of affected by Uterine contractions, Light decelerations, Severe decelerations,,Prolongued decelerations, Abnormal short term variability., Mean value of short term variability, Histogram mode
compared to the other two clusters. It also has higher levels affected by Histogram mean, Histogram median, Histogram variance.
In order to validate the clusters, ANOVA analysis was conducted to test for significant differences between the clusters on fetal health. Results indicated significant differences between the clusters on GPA (F(2, 1485)= 420.2, p<.0001). The tukey test showed that clusters differ significantly within fetal health, although difference between cluster 2 and 3 is not significant.
CODE:
!pip install statsmodels
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
from scipy.spatial.distance import cdist
from sklearn.decomposition import PCA
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
%matplotlib inline
RND_STATE = 2226
!pip install pandas
!pip install numpy
!pip install matplotlib
!pip install statsmodels
data = pd.read_csv("fetal_health.csv")
data.columns = map(str.upper, data.columns)
data_clean = data.dropna()
AH_data = pd.read_csv("fetal_health.csv")
data_clean = AH_data.dropna()
cluster=data_clean[['baseline value','uterine_contractions','light_decelerations','severe_decelerations',
'prolongued_decelerations',
'abnormal_short_term_variability','mean_value_of_short_term_variability',
'histogram_mode','histogram_mean',
'histogram_median','histogram_variance']]
cluster.describe()
clustervar=cluster.copy()
clustervar['baseline value']=preprocessing.scale(clustervar['baseline value'].astype('float64'))
clustervar['uterine_contractions']=preprocessing.scale(clustervar['uterine_contractions'].astype('float64'))
clustervar['light_decelerations']=preprocessing.scale(clustervar['light_decelerations'].astype('float64'))
clustervar['severe_decelerations']=preprocessing.scale(clustervar['severe_decelerations'].astype('float64'))
clustervar['prolongued_decelerations']=preprocessing.scale(clustervar['prolongued_decelerations'].astype('float64'))
clustervar['abnormal_short_term_variability']=preprocessing.scale(clustervar['abnormal_short_term_variability'].astype('float64'))
clustervar['mean_value_of_short_term_variability']=preprocessing.scale(clustervar['mean_value_of_short_term_variability'].astype('float64'))
clustervar['histogram_mode']=preprocessing.scale(clustervar['histogram_mode'].astype('float64'))
clustervar['histogram_mean']=preprocessing.scale(clustervar['histogram_mean'].astype('float64'))
clustervar['histogram_median']=preprocessing.scale(clustervar['histogram_median'].astype('float64'))
clustervar['histogram_variance']=preprocessing.scale(clustervar['histogram_variance'].astype('float64'))
clus_train, clus_test = train_test_split(clustervar, test_size=0.3, random_state=RND_STATE)
!pip install cluster
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
!pip install statsmodels
!pip install matplotlib
!pip install scipy
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
plt.show()
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
clus_train.reset_index(level=0, inplace=True)
cluslist=list(clus_train['index'])
labels=list(model3.labels_)
newlist=dict(zip(cluslist, labels))
newclus=DataFrame.from_dict(newlist, orient='index')
newclus.columns = ['cluster']
newclus.describe()
newclus.reset_index(level=0, inplace=True)
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
merged_train.cluster.value_counts()
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
gpa_data=data_clean['fetal_health']
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=RND_STATE)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['fetal_health', 'cluster']].dropna()
gpamod = smf.ols(formula='fetal_health ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for fetal health by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for fetal health by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['fetal_health'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
0 notes
Text
HW.3 : Lasso Regression
At this HW. I have implemented lasso regression to predict Fetal Health on a list of explanatory variables. This time I’ve also selected all variables, that exist in given dataset. (baseline value, accelerations, fetal movement, uterine contractions, light decelerations, severe decelerations, prolongued decelerations, etc). All of them were used to build final model to predict the dependent variable – fetal_health .
In order to fit the Lasso Regression model, which helps to improve overall model quality and removes unimportant variables by adding an additional coefficient – alpha to each explanatory variable, I had to perform some preprocessing on data. In addiction to usual procedure of incomplete data removal, I’ve also added scaling to all of variables in order to lead it to one dimension.
In order to test final model I’ve split data into two sets – train (70%) and test(30%) to train and test Lasso Regression model respectively. Moreover, to reduce the influence of data imbalance I’ve added cv parameter (cv=10, default is 3) in order to specify the number of folds in a Stratified KFold, which helps to solve this problem. Change in the validation mean square error at each step:

in[19]: pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, target, test_size=.3, random_state=RND_STATE)
Applying the Lasso Regression to the data assigns a Regression Coefficient to each predictor. Predictors with a Regression Coefficient of zero were eliminated,18 were retained.
{'baseline value': 0.18743121710692548 'accelerations': -0.012420438629578371, 'fetal_movement': -0.02678008761229712, 'uterine_contractions': -0.08881958546068254, 'light_decelerations': -0.025262653640074875, 'severe_decelerations': 0.0362727278281784, 'prolongued_decelerations': 0.20629467915907235, 'mean_value_of_short_term_variability': -0.034597706276368205, 'percentage_of_time_with_abnormal_long_term_variability': 0.2430909703179747, 'mean_value_of_long_term_variability': 0.0, 'histogram_width': 0.0, 'histogram_min': 0.08839831243779736, 'histogram_max': 0.05884828569739267, 'histogram_number_of_peaks': 0.0, 'histogram_number_of_zeroes': 0.0009254817992133355, 'histogram_mode': -0.10936885075312966, 'histogram_mean': -0.19033727038108944, 'histogram_median': 0.0, 'histogram_variance': 0.0865905038625272, 'histogram_tendency': 0.045210673662552284}
As seen, there 4 variables –mean value of long term variability, histogram width,histogram number of peaks, histogram median were removed by algorithm. It can be explained with the fact that Lasso Regression removes correlating variables from dataset.
The MSE for the training data stood at 0.1558 while it was 0.1716 for the test data.
IN[26] : train_error = mean_squared_error(tar_train, model.predict(pred_train))
test_error = mean_squared_error(tar_test, model.predict(pred_test))
print('training data MSE', train_error)
print('test data MSE', test_error)
training data MSE 0.1558480179256629 test data MSE 0.17166571196183492
Regression Coefficients Progression for Lasso Paths:

CODE:
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LassoLarsCV
from sklearn import preprocessing
from sklearn.metrics import mean_squared_error
pd.options.mode.chained_assignment = None
%matplotlib inline
RND_STATE = 2226
data = pd.read_csv("fetal_health.csv") data.columns = map(str.upper, data.columns) data.describe()
data_clean = data.dropna()
AH_data = pd.read_csv("fetal_health.csv")
data_clean = AH_data.dropna()
predvar = data_clean[['baseline value','accelerations','fetal_movement','uterine_contractions','light_decelerations','severe_decelerations','prolongued_decelerations','mean_value_of_short_term_variability', 'percentage_of_time_with_abnormal_long_term_variability','mean_value_of_long_term_variability','histogram_width','histogram_min','histogram_max','histogram_number_of_peaks','histogram_number_of_zeroes','histogram_mode','histogram_mean','histogram_median','histogram_variance','histogram_tendency']] target = data_clean.fetal_health
recode = {1:1, 2:0} data_clean['baseline value'] = data_clean['baseline value'].map(recode)
predictors=predvar.copy()
predictors['baseline value']=preprocessing.scale(predictors['baseline value'].astype('float64'))
predictors['accelerations']=preprocessing.scale(predictors['accelerations'].astype('float64'))
predictors['uterine_contractions']=preprocessing.scale(predictors['uterine_contractions'].astype('float64'))
predictors['light_decelerations']=preprocessing.scale(predictors['light_decelerations'].astype('float64'))
predictors['severe_decelerations']=preprocessing.scale(predictors['severe_decelerations'].astype('float64'))
predictors['prolongued_decelerations']=preprocessing.scale(predictors['prolongued_decelerations'].astype('float64'))
predictors['mean_value_of_short_term_variability']=preprocessing.scale(predictors['mean_value_of_short_term_variability'].astype('float64'))
predictors['percentage_of_time_with_abnormal_long_term_variability']=preprocessing.scale(predictors['percentage_of_time_with_abnormal_long_term_variability'].astype('float64'))
predictors['mean_value_of_long_term_variability']=preprocessing.scale(predictors['mean_value_of_long_term_variability'].astype('float64'))
predictors['histogram_width']=preprocessing.scale(predictors['histogram_width'].astype('float64'))
predictors['histogram_min']=preprocessing.scale(predictors['histogram_min'].astype('float64'))
predictors['histogram_max']=preprocessing.scale(predictors['histogram_max'].astype('float64'))
predictors['histogram_number_of_peaks']=preprocessing.scale(predictors['histogram_number_of_peaks'].astype('float64'))
predictors['histogram_number_of_zeroes']=preprocessing.scale(predictors['histogram_number_of_zeroes'].astype('float64'))
predictors['histogram_mode']=preprocessing.scale(predictors['histogram_mode'].astype('float64'))
predictors['histogram_mean']=preprocessing.scale(predictors['histogram_mean'].astype('float64'))
predictors['histogram_median']=preprocessing.scale(predictors['histogram_median'].astype('float64'))
predictors['histogram_variance']=preprocessing.scale(predictors['histogram_variance'].astype('float64'))
predictors['histogram_tendency']=preprocessing.scale(predictors['histogram_tendency'].astype('float64'))
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, target, test_size=.3, random_state=RND_STATE)
model = LassoLarsCV(cv=10, precompute=False).fit(pred_train,tar_train)
dict(zip(predictors.columns, model.coef_))
m_log_alphas = -np.log10(model.alphas_)
ax = plt.gca()
plt.plot(m_log_alphas, model.coef_path_.T)
plt.axvline(-np.log10(model.alpha_), linestyle='--', color='k',
label='alpha CV')
plt.ylabel('Regression Coefficients')
plt.xlabel('-log(alpha)')
plt.title('Regression Coefficients Progression for Lasso Paths')
m_log_alphascv = -np.log10(model.cv_alphas_)
plt.figure()
plt.plot(m_log_alphascv, model.mse_path_, ':')
plt.plot(m_log_alphascv, model.mse_path_.mean(axis=-1), 'k',
label='Average across the folds', linewidth=2)
plt.axvline(-np.log10(model.alpha_), linestyle='--', color='k',
label='alpha CV')
plt.legend()
plt.xlabel('-log(alpha)')
plt.ylabel('Mean squared error')
plt.title('Mean squared error on each fold')
train_error = mean_squared_error(tar_train, model.predict(pred_train))
test_error = mean_squared_error(tar_test, model.predict(pred_test))
print('training data MSE', train_error)
print('test data MSE', test_error)
rsquared_train=model.score(pred_train,tar_train)
rsquared_test=model.score(pred_test,tar_test)
print('training data R-square', rsquared_train)
print('test data R-square', rsquared_test)
0 notes
Text
Fetal Health Classification(Assignment2-Running a Random Forest)
Task:
Run a Random Forest.
You will need to perform a random forest analysis to evaluate the importance of a series of explanatory variables in predicting a binary, categorical response variable.
Solution:
In assignment No. 1, we chose four variables, which are Fetal movement ,Uterine contractions ,Histogram number of peaks and Histogram number of zeros to study the extent of their impact on the health of the fetus. In this assignment, we wrote a code that makes a random comparison between all the factors in the datasets that we applied in the first assignment. We obtained completely different results from what we obtained in the first assignment. The factors that are considered indicators of the health of the fetus here were other than those we chose in the previous assignment. As for the accuracy that we obtained here, it was greatly excellent than it was in the previous assignment, as we got here an accuracy of 93.5%, but in the previous assignment it was 74%.
Reading data from file

To show the frame of datasets:

Apply the Random Forest in Python:
Split into train test datasets
Split into training and testing sets
predictors = data_clean[['baseline value','accelerations','fetal_movement', 'uterine_contractions', 'light_decelerations', 'severe_decelerations', 'prolongued_decelerations',
'abnormal_short_term_variability', 'mean_value_of_short_term_variability', 'percentage_of_time_with_abnormal_long_term_variability','mean_value_of_long_term_variability','histogram_width','histogram_min','histogram_max','histogram_number_of_peaks','histogram_number_of_zeroes','histogram_mode',
'histogram_mean',
'histogram_median','histogram_variance', 'histogram_tendency']]
targets = data_clean.fetal_health
Apply train_test_split.
For example, you can set the test size to 0.4, and therefore the model testing will be based on 40% of the dataset, while the model training will be based on 60% of the dataset:
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, targets, test_size=.4, random_state=RND_STATE)
Build a Random Forest Classifier
Fitting RandomForestClassifier
Apply the Random Forest as follows:
classifier = RandomForestClassifier(n_estimators=22, random_state=RND_STATE)
classifier = classifier.fit(pred_train, tar_train)
predictions = classifier.predict(pred_test)
print the Accuracy
print(confusion_matrix(tar_test, predictions))
print()
print("Accuracy: ", accuracy_score(tar_test, predictions))
The output
Confusion matrix:
[[642 10 4]
[ 28 94 5]
[ 3 5 60]]
Accuracy: 0.9353701527614571
Final model looked excellent on test data and showed accuracy level at 93.5%!
Find important features with Random Forest model
important_features = pd.Series(data=classifier.feature_importances_,index=predictors.columns) important_features.sort_values(ascending=False,inplace=True)
After fitting the model it occurred that these factors influence final variable with different level of importance. So, I’ve calculated and sorted descending these factors into a feature importance list:
abnormal_short_term_variability 0.151485
percentage_of_time_with_abnormal_long_term_variability 0.118127
mean_value_of_short_term_variability 0.090103
histogram_mean 0.080043
prolongued_decelerations 0.076003
histogram_median 0.065495
histogram_mode 0.054925
accelerations 0.044466
uterine_contractions 0.044239
baseline value 0.042262
mean_value_of_long_term_variability 0.037475
histogram_variance 0.034731
histogram_min 0.034260
histogram_width 0.033916
histogram_max 0.028461
histogram_number_of_peaks 0.025365
fetal_movement 0.018233
light_decelerations 0.008666
histogram_number_of_zeroes 0.006063
histogram_tendency 0.005682
severe_decelerations 0.000000
dtype: float64
we see that the more factors that influnces the Fetal health
are:
abnormal_short_term_variability
percentage_of_time_with_abnormal_long_term_variability
mean_value_of_short_term_variability
histogram_mean
prolongued_decelerations
histogram_median
tested how number of trees in random forest influences on final model accuracy. So results can be presented in this plot:

As it shows, even one tree is able to show accuracy at a excellent level. So, this data can be described even with one tree. But, on the other hand, it is clear, that after adding some more trees (>7) final accuracy increases a bit, making model able to predict data in a better way.
Python code:
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
%matplotlib inline
RND_STATE = 2226
AH_data = pd.read_csv("fetal_health.csv")
data_clean = AH_data.dropna()
data_clean.dtypes
data_clean.describe()
predictors = data_clean[['baseline value','accelerations','fetal_movement', 'uterine_contractions', 'light_decelerations', 'severe_decelerations', 'prolongued_decelerations',
'abnormal_short_term_variability', 'mean_value_of_short_term_variability', 'percentage_of_time_with_abnormal_long_term_variability','mean_value_of_long_term_variability','histogram_width','histogram_min','histogram_max','histogram_number_of_peaks','histogram_number_of_zeroes','histogram_mode',
'histogram_mean',
'histogram_median','histogram_variance', 'histogram_tendency']]
targets = data_clean.fetal_health
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, targets, test_size=.4, random_state=RND_STATE)
print("Predict train shape: ", pred_train.shape)
print("Predict test shape: ", pred_test.shape)
print("Target train shape: ", tar_train.shape)
print("Target test shape: ", tar_test.shape)
classifier = RandomForestClassifier(n_estimators=22, random_state=RND_STATE)
classifier = classifier.fit(pred_train, tar_train)
predictions = classifier.predict(pred_test)
print("Confusion matrix:")
print(confusion_matrix(tar_test, predictions))
print()
print("Accuracy: ", accuracy_score(tar_test, predictions))
important_features = pd.Series(data=classifier.feature_importances_,index=predictors.columns)
important_features.sort_values(ascending=False,inplace=True)
important_features
model = ExtraTreesClassifier(random_state=RND_STATE)
model.fit(pred_train, tar_train)
print(model.feature_importances_)
trees = range(22)
accuracy = np.zeros(22)
for idx in range(len(trees)):
classifier = RandomForestClassifier(n_estimators=idx + 1, random_state=RND_STATE)
classifier = classifier.fit(pred_train, tar_train)
predictions = classifier.predict(pred_test)
accuracy[idx] = accuracy_score(tar_test, predictions)
plt.cla()
plt.plot(trees, accuracy)
plt.show()
0 notes
Text
Fetal Health Classification (Decision Trees Assigment 1)
Task:
This week’s assignment involves decision trees, and more specifically, classification trees. Decision trees are predictive models that allow for a data driven exploration of nonlinear relationships and interactions among many explanatory variables in predicting a response or target variable. When the response variable is categorical (two levels), the model is a called a classification tree. Explanatory variables can be either quantitative, categorical or both. Decision trees create segmentations or subgroups in the data, by applying a series of simple rules or criteria over and over again which choose variable constellations that best predict the response (i.e. target) variable.
Run a Classification Tree.
You will need to perform a decision tree analysis to test nonlinear relationships among a series of explanatory variables and a binary, categorical response variable.
Generated decision tree :

OPEN IMAGE
2126 measurements extracted from cardiotocograms and classified by expert obstetricians.
Decision tree analysis was performed to test nonlinear relationships among a series of explanatory variables and a binary, categorical response variable. All possible separations (categorical) or cut points (quantitative) are tested.
My decision tree uses these variables to predict output variable (fetal_health) – whether fetal health is good or no:
Fetal movement (fetal_movement=flMO).
Uterine contractions (uterin_contractions=Utrin).
Histogram number of peaks (histomgram_number_of_peaks=Peak).
Histogram number of zeros ( histomgram_number_of_ zeros=Zero).
After fitting the tree, I’ve tested it on test dataset and got accuracy = 0.74. This is a good result for a model, which is based only on four explaining variables.
From decision tree we can observe:
Uterine contractions are the most important factor to determine if the fetal is in a good or no.
When Fetal movement is large value it means that the fetal is in good state.
When Histogram number of peaks are high mean that fetal is in good condition
Formatted Source code (and output)
!pip install pydotplus
import pandas as pd import sklearn.metrics from numpy.lib.format import magic from sklearn.model_selection import train_test_split from sklearn.tree import DecisionTreeClassifier from sklearn import tree from io import StringIO from IPython.display import Image import pydotplus RND_STATE =2226
AH_data = pd.read_csv("fetal_health.csv") data_clean = AH_data.dropna() data_clean.dtypes data_clean.describe()

predictors = data_clean[['fetal_movement','uterine_contractions', 'histogram_number_of_peaks', 'histogram_number_of_zeroes']]
targets = data_clean.fetal_health
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, targets, test_size=0.3)
classifier=DecisionTreeClassifier(random_state=RND_STATE)
classifier=classifier.fit(pred_train, tar_train) predictions=classifier.predict(pred_test)
print("Confusion matrix:\n", sklearn.metrics.confusion_matrix(tar_test,predictions)) print("Accuracy: ",sklearn.metrics.accuracy_score(tar_test, predictions))

out = StringIO() tree.export_graphviz(classifier, out_file=out, feature_names=["flMo", "Utrin", "Peak","Zero"],proportion=True, filled=True, max_depth=4) graph=pydotplus.graph_from_dot_data(out.getvalue()) img = Image(data=graph.create_png()) img
with open("utput" + ".png", "wb") as f: f.write(img.data)
0 notes