Don't wanna be here? Send us removal request.
Text
varianza Anova
import numpy
import pandas
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
data = pandas.read_csv('nesarc.csv', low_memory=False)
#setting variables you will be working with to numeric
data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1'], errors='coerce')
data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1'], errors='coerce')
data['CHECK321'] = pandas.to_numeric(data['CHECK321'], errors='coerce')
#subset data to young adults age 18 to 25 who have smoked in the past 12 months
sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
#SETTING MISSING DATA
sub1['S3AQ3B1']=sub1['S3AQ3B1'].replace(9, numpy.nan)
sub1['S3AQ3C1']=sub1['S3AQ3C1'].replace(99, numpy.nan)
#recoding number of days smoked in the past month
recode1 = {1: 30, 2: 22, 3: 14, 4: 5, 5: 2.5, 6: 1}
sub1['USFREQMO']=sub1['S3AQ3B1'].map(recode1)
#converting new variable USFREQMMO to numeric
sub1['USFREQMO']=pandas.to_numeric(sub1['USFREQMO'], errors='coerce')
# Creating a secondary variable multiplying the days smoked/month and the number of cig/per day
sub1['NUMCIGMO_EST']=sub1['USFREQMO'] *sub1['S3AQ3C1']
sub1['NUMCIGMO_EST']=pandas.to_numeric(sub1['NUMCIGMO_EST'], errors='coerce')
ct1 =sub1.groupby('NUMCIGMO_EST').size()
print (ct1)
# using ols function for calculating the F-statistic and associated p value
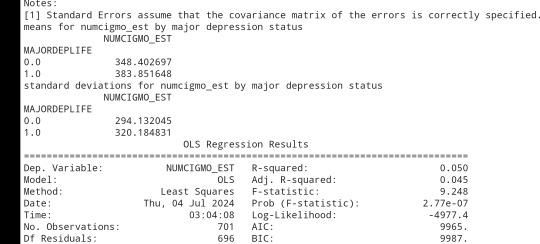
model1 =smf.ols(formula='NUMCIGMO_EST ~ C(MAJORDEPLIFE)', data=sub1)
results1 = model1.fit()
print (results1.summary())
sub2 = sub1[['NUMCIGMO_EST', 'MAJORDEPLIFE']].dropna()
print ('means for numcigmo_est by major depression status')
m1=sub2.groupby('MAJORDEPLIFE').mean()
print (m1)
print ('standard deviations for numcigmo_est by major depression status')
sd1 =sub2.groupby('MAJORDEPLIFE').std()print (sd1)
#i will call it sub3
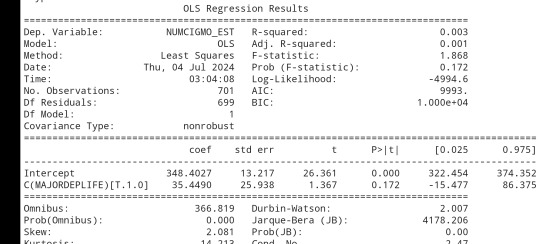
sub3 = sub1[['NUMCIGMO_EST', 'ETHRACE2A']].dropna()
model2 =smf.ols(formula='NUMCIGMO_EST ~ C(ETHRACE2A)', data=sub3).fit()
print (model2.summary())
print ('means for numcigmo_est by major depression status')
m2=sub3.groupby('ETHRACE2A').mean()
print (m2)
print ('standard deviations for numcigmo_est by major depression status')
sd2=sub3.groupby('ETHRACE2A').std()print (sd2)
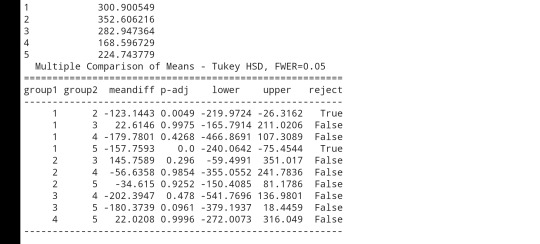
mc1=multi.MultiComparison(sub3['NUMCIGMO_EST'], sub3['ETHRACE2A'])
res1 = mc1.tukeyhsd()
print(res1.summary())


0 notes
Text
K-means clusthering
#importamos las librerías necesarias
from pandas import Series,DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection
import train_test_splitfrom sklearn
import preprocessing
from sklearn.cluster import KMeans
"""Data Management"""
data =pd.read_csv("/content/drive/MyDrive/tree_addhealth.csv" )
#upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
# Data Managementdata_clean = data.dropna()
# subset clustering variablescluster=data_clean[['ALCEVR1','MAREVER1','ALCPROBS1','DEVIANT1','VIOL1','DEP1','ESTEEM1','SCHCONN1','PARACTV', 'PARPRES','FAMCONCT']]
cluster.describe()
# standardize clustering variables to have mean=0 and sd=1
clustervar=cluster.copy()
clustervar['ALCEVR1']=preprocessing.scale(clustervar['ALCEVR1'].astype('float64'))
clustervar['ALCPROBS1']=preprocessing.scale(clustervar['ALCPROBS1'].astype('float64'))
clustervar['MAREVER1']=preprocessing.scale(clustervar['MAREVER1'].astype('float64'))
clustervar['DEP1']=preprocessing.scale(clustervar['DEP1'].astype('float64'))
clustervar['ESTEEM1']=preprocessing.scale(clustervar['ESTEEM1'].astype('float64'))
clustervar['VIOL1']=preprocessing.scale(clustervar['VIOL1'].astype('float64'))
clustervar['DEVIANT1']=preprocessing.scale(clustervar['DEVIANT1'].astype('float64'))
clustervar['FAMCONCT']=preprocessing.scale(clustervar['FAMCONCT'].astype('float64'))
clustervar['SCHCONN1']=preprocessing.scale(clustervar['SCHCONN1'].astype('float64'))
clustervar['PARACTV']=preprocessing.scale(clustervar['PARACTV'].astype('float64'))
clustervar['PARPRES']=preprocessing.scale(clustervar['PARPRES'].astype('float64'))
# split data into train and test sets
clus_train, clus_test = train_test_split(clustervar, test_size=.3, random_state=123)
# k-means cluster analysis for 1-9 clusters
from scipy.spatial.distance import cdist
clusters=range(1,10)
meandist=[]
for k in clusters: model=KMeans(n_clusters=k) model.fit(clus_train) clusassign=model.predict(clus_train) meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
"""Plot average distance from observations from the cluster centroidto use the Elbow Method to identify number of clusters to choose"""
plt.plot(clusters,meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
# Interpret 2 cluster solution
model2=KMeans(n_clusters=2)model2.fit(clus_train)
clusassign=model2.predict(clus_train)
# plot clusters
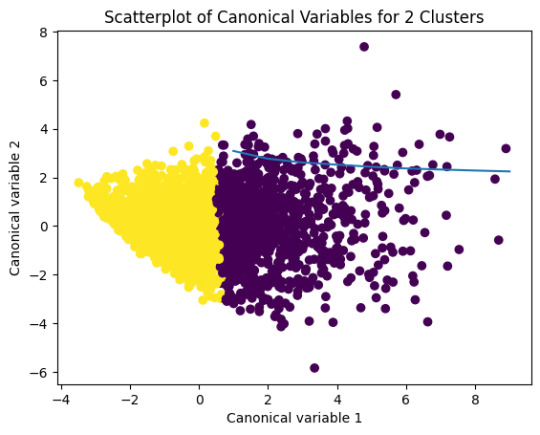
from sklearn.decomposition import PCA
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0],y=plot_columns[:,1],c=model2.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 2 Clusters')
plt.show()
"""BEGIN multiple steps to merge cluster assignment with clustering variables to examinecluster variable means by cluster"""
# create a unique identifier variable from the index for the
# cluster training data to merge with the cluster assignment variable
clus_train.reset_index(level=0, inplace=True)
# create a list that has the new index variable
cluslist=list(clus_train['index'])
# create a list of cluster assignments
labels=list(model2.labels_)
# combine index variable list with cluster assignment list into a dictionary
newlist=dict(zip(cluslist, labels))newlist
# convert newlist dictionary to a dataframenew
clus=DataFrame.from_dict(newlist, orient='index')newclus
# rename the cluster assignment columnnew
clus.columns = ['cluster']
# now do the same for the cluster assignment variable
# create a unique identifier variable from the index for the
# cluster assignment dataframe
# to merge with cluster training datanew
clus.reset_index(level=0, inplace=True)
# merge the cluster assignment dataframe with the cluster training variable dataframe
# by the index variablemerged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
# cluster frequenciesmerged_train.cluster.value_counts()
"""END multiple steps to merge cluster assignment with clustering variables to examinecluster variable means by cluster"""
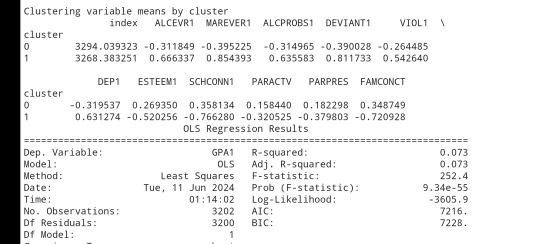
# FINALLY calculate clustering variable means by
clusterclustergrp = merged_train.groupby('cluster').mean()print ("Clustering variable means by cluster")
print(clustergrp)
# validate clusters in training data by examining cluster differences in GPA using ANOVA
# first have to merge GPA with clustering variables and cluster assignment data
gpa_data=data_clean['GPA1']
# split GPA data into train and test sets
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=123)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['GPA1', 'cluster']].dropna()
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
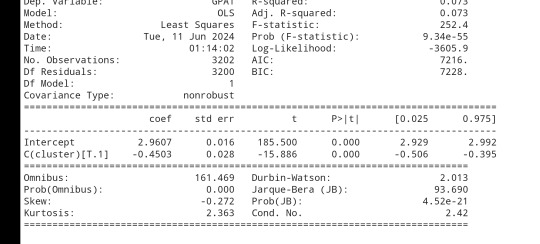
gpamod = smf.ols(formula='GPA1 ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
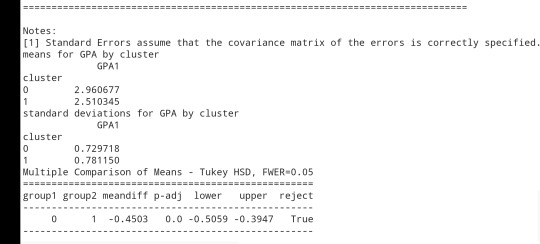
print ('means for GPA by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()print (m2)
mc1 = multi.MultiComparison(sub1['GPA1'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
0 notes
Text
Regresión LassoLarscv
Primeramente cargamos las librerías necesarias:
#from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LassoLarsCV
#Load the dataset
#Nota: al cargar los datos, es recomendable cargar los datos en el mismo entorno de ejecución, en caso de usar google.colab, puede conectar su drive y posteriormente copiar la ruta del archivo de "tree_addhealth.csv"
#Ejemplo: df=pd. read_csv('/content/drive/MyDrive/tree_addhealth.csv' )
#Continuando leemos los datos:
data = pd.read_csv("tree_addhealth.csv")
#upper-case all DataFrame column names
data.columns = map(str.upper, data.columns)
# Data Management
data_clean = data.dropna()
recode1 = {1:1, 2:0}
data_clean['MALE']=data_clean['BIO_SEX'].map(recode1)
#select predictor variables and target variable as separate data sets
predvar=data_clean[['MALE','HISPANIC','WHITE','BLACK','NAMERICAN','ASIAN','AGE','ALCEVR1','ALCPROBS1','MAREVER1','COCEVER1','INHEVER1','CIGAVAIL','DEP1','ESTEEM1','VIOL1','PASSIST','DEVIANT1','GPA1','EXPEL1','FAMCONCT','PARACTV','PARPRES']]
target = data_clean.SCHCONN1
# standardize predictors to have mean=0 and sd=1
predictors=predvar.copy()
from sklearn import preprocessing
predictors['MALE']=preprocessing.scale(predictors['MALE'].astype('float64'))
predictors['HISPANIC']=preprocessing.scale(predictors['HISPANIC'].astype('float64'))
predictors['WHITE']=preprocessing.scale(predictors['WHITE'].astype('float64'))
predictors['NAMERICAN']=preprocessing.scale(predictors['NAMERICAN'].astype('float64'))
predictors['ASIAN']=preprocessing.scale(predictors['ASIAN'].astype('float64'))
predictors['AGE']=preprocessing.scale(predictors['AGE'].astype('float64'))
predictors['ALCEVR1']=preprocessing.scale(predictors['ALCEVR1'].astype('float64'))
predictors['ALCPROBS1']=preprocessing.scale(predictors['ALCPROBS1'].astype('float64'))
predictors['MAREVER1']=preprocessing.scale(predictors['MAREVER1'].astype('float64'))
predictors['COCEVER1']=preprocessing.scale(predictors['COCEVER1'].astype('float64'))
predictors['INHEVER1']=preprocessing.scale(predictors['INHEVER1'].astype('float64'))
predictors['CIGAVAIL']=preprocessing.scale(predictors['CIGAVAIL'].astype('float64'))
predictors['DEP1']=preprocessing.scale(predictors['DEP1'].astype('float64'))
predictors['ESTEEM1']=preprocessing.scale(predictors['ESTEEM1'].astype('float64'))
predictors['VIOL1']=preprocessing.scale(predictors['VIOL1'].astype('float64'))
predictors['PASSIST']=preprocessing.scale(predictors['PASSIST'].astype('float64'))
predictors['DEVIANT1']=preprocessing.scale(predictors['DEVIANT1'].astype('float64'))
predictors['GPA1']=preprocessing.scale(predictors['GPA1'].astype('float64'))
predictors['EXPEL1']=preprocessing.scale(predictors['EXPEL1'].astype('float64'))
predictors['FAMCONCT']=preprocessing.scale(predictors['FAMCONCT'].astype('float64'))
predictors['PARACTV']=preprocessing.scale(predictors['PARACTV'].astype('float64'))
predictors['PARPRES']=preprocessing.scale(predictors['PARPRES'].astype('float64'))
#split data into train and test sets
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, target,test_size=.3, random_state=123)
# specify the lasso regression model
model=LassoLarsCV(cv=10, precompute=False).fit(pred_train,tar_train)
# print variable names and regression coefficients
dict(zip(predictors.columns, model.coef_))

#Como podemos notar, las variables: 'PARPRES', 'PASSIST', 'INHEVER1' 'ALCPROBS1' no influyen en la predicción, pues sus coeficientes se van a 0 cuando se realiza la regularización Lasso, por tanto se eliminan del modelo final.
De aquí igual podemos ver que:
Las variables de ESTEEM1 y DEP1 tuvieron los coeficientes más altos con 1.1 y -0.86,respectivamente, estas variables de autoestima( ESTEEM1) y depresión(DEP1) se asocian más fuerte a la conectividad escolar (SCHCONN1), posteriormente se relaciona las variables BLACK con - 0.69, GPA1 con 0.67 y VIOL1 con - 0.64.
De las cuales Autoestima y GPA1 se asociaron positivamente a nuestra target (SCHCONN1), y la depresión, la etnia negra (BLACK) y VIOL1 se asociaron negativamente a nuestra target
# plot coefficient progression
m_log_alphas = -np.log10(model.alphas_)
ax = plt.gca()
plt.plot(m_log_alphas, model.coef_path_.T)
plt.axvline(-np.log10(model.alpha_), linestyle='--', color='k', label='alpha CV')
plt.ylabel('Regression Coefficients')
plt.xlabel('-log(alpha)')
plt.title('Regression Coefficients Progression for Lasso Paths')

# plot mean square error for each fold
m_log_alphascv = - np.log10(model.cv_alphas_)
# specify the lasso regression model
model=LassoLarsCV(cv=10, precompute=False).fit(pred_train,tar_train)plt.figure()
plt.plot(m_log_alphascv, model.mse_path_, ':')
plt.plot(m_log_alphascv, model.mse_path_.mean(axis=-1), 'k', label='Average across the folds', linewidth=2)
plt.axvline(-np.log10(model.alpha_), linestyle='--', color='k', label='alpha CV')
plt.legend()
plt.xlabel('-log(alpha)')
plt.ylabel('Mean squared error')
plt.title('Mean squared error on each fold')

# MSE from training and test data
from sklearn.metrics import mean_squared_error
train_error = mean_squared_error(tar_train, model.predict(pred_train))
test_error = mean_squared_error(tar_test, model.predict(pred_test))
print ('training data MSE')
print(train_error)
print ('test data MSE')
print(test_error)

Esto sugiere que la precisión de la predicción fue bastante estable en entre el training data MSE y el test data MSE, y también tenemos que fué menos preciso al predecir la conectividad escolar con el test data
# R-square from training and test data
rsquared_train=model.score(pred_train,tar_train)
rsquared_test=model.score(pred_test,tar_test)
print ('training data R-square')
print(rsquared_train)
print ('test data R-square')
print(rsquared_test)

Los valores de R cuadrada, significan que el modelo que tenemos explicó el 33% y el 31% de la varianza en la conectividad escolar en los conjuntos de entrenamiento(training data) y de prueba(test data) , respectivamente.
0 notes
Text
Bosques aleatorios
Importamos librerías necesarias
from pandas import Series, DataFrameimport pandas as pd
import numpy as np
import os
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import classification_report
import sklearn.metrics
# Feature Importance
from sklearn import datasets
from sklearn.ensemble import ExtraTreesClassifier
A continuación cargamos los datos a realizar el análisis exploratorio, en mi caso cargué los datos de forma local, así recomiendo cargar los datos adaptado a su directorio adaptando solo '/content/drive/MyDrive' especificando en la ruta en la que se encuentra
# Ruta base donde se encuentra tu archivo
base_path = '/content/drive/MyDrive'
# Nombre de tu archivo
file_name = 'tree_addhealth.csv'
# Usando os.path.join() para unir la ruta base y el nombre del archivo
full_path = os.path.join(base_path, file_name)
#Load the dataset
AH_data = pd.read_csv(full_path)
data_clean = AH_data.dropna()
data_clean.dtypes
data_clean.describe()

Seleccionamos las variables predictoras, con base en el análisis exploratorio y demás:
#Split into training and testing setspredictors = data_clean[['BIO_SEX','HISPANIC','WHITE','BLACK','NAMERICAN','ASIAN','age','ALCEVR1','ALCPROBS1','marever1','cocever1','inhever1','cigavail','DEP1','ESTEEM1','VIOL1','PASSIST','DEVIANT1','SCHCONN1','GPA1','EXPEL1','FAMCONCT','PARACTV','PARPRES']]
targets = data_clean.TREG1
pred_train, pred_test, tar_train, tar_test = train_test_split(predictors, targets, test_size=.4)
pred_train.shape

Éstos son los tamaños respectivos de cada variable, predictora y Target, que se toman.
#Build model on training datafrom sklearn.ensemble import RandomForestClassifier
classifier=RandomForestClassifier(n_estimators=25)
classifier=classifier.fit(pred_train,tar_train)
predictions=classifier.predict(pred_test)
sklearn.metrics.confusion_matrix(tar_test,predictions)
sklearn.metrics.accuracy_score(tar_test, predictions)
Out: 0.8366120218579235
# fit an Extra Trees model to the data
model =ExtraTreesClassifier()
model.fit(pred_train,tar_train)
# display the relative importance of each attribute
print(model.feature_importances_)

donde la más alta es de 0.11411386, que es el valor de la variable de consumo de cannabis.
"""Running a different number of trees and see the effect of that on the accuracy of the prediction"""
trees=range(25)
accuracy=np.zeros(25)
for idx in range(len(trees)): classifier=RandomForestClassifier(n_estimators=idx + 1) classifier=classifier.fit(pred_train,tar_train) predictions=classifier.predict(pred_test) accuracy[idx]=sklearn.metrics.accuracy_score(tar_test, predictions)
plt.cla()
plt.plot(trees, accuracy)

Como podemos ver en el bosque, para el primer valor o árbol, tenemos que tiene una precisión bastante buena, casi del 83% y si vemos este varia de entre 84% y 79% de precision, con forme avanza y se analizan más árboles, también tenemos que nuesto modelo en particular tiene una precisión de 0.8366120218579235, es decir, que clasifica con una certeza del 83% entre fumadores habituales y no habituales, ésto pues era la pregunta que nos hicimos al principio para hacer la clasificación
"¿En los últimos 30 días consumiste un cigarrillo?", también por la impresión de "model.feature_importances_", vimos que la variable de consumo de marihuana, puede ayudar a contestar mejor a dicha pregunta o entenderla.
0 notes
Text
árbol de clasificación
import matplotlib.pylab as pltfrom sklearn.tree import plot_treefrom sklearn.datasets import load_irisfrom sklearn.model_selection import train_test_splitfrom sklearn.tree import DecisionTreeClassifierfrom sklearn.metrics import accuracy_score
# Cargar un conjunto de datos como ejemploiris = load_iris()
X, y = iris.data, iris.target
# Dividir los datos en conjuntos de entrenamiento y prueba
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=1)
# Crear el modelo de árbol de decisión
clf = DecisionTreeClassifier()
# Entrenar el modeloclf.fit(X_train, y_train)
# Predecir las respuestas para el conjunto de datos de prueba
y_pred = clf.predict(X_test)
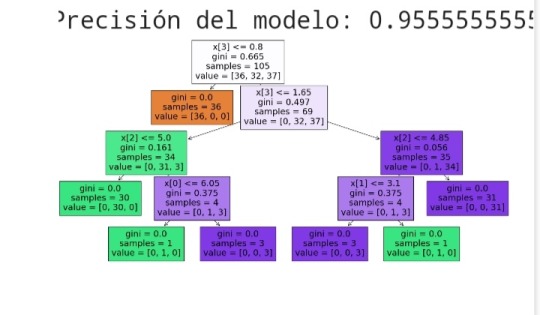
# Calcular la precisión del modelo
accuracy = accuracy_score(y_test, y_pred)
print(f'Precisión del modelo: {accuracy}')
plt.figure(figsize=(20,10))
plot_tree(clf, filled=True)
plt.show()
1 note
·
View note