Don't wanna be here? Send us removal request.
Text
Assignment 4 - Data Visualization
import pandas import numpy import seaborn import matplotlib.pyplot as plt
data = pandas.read_csv('gapminder.csv', low_memory=False)
data = data.replace(r'^\s*$', numpy.NaN, regex=True)
#setting variables you will be working with to numeric
data['internetuserate'] = pandas.to_numeric(data['internetuserate']) data['urbanrate'] = pandas.to_numeric(data['urbanrate']) data['incomeperperson'] = pandas.to_numeric(data['incomeperperson']) data['hivrate'] = pandas.to_numeric(data['hivrate'])
desc1 = data['urbanrate'].describe()
#The below are descriptive statistics results for all the 2 variables used in the code
print (desc1)
count 203.000000
mean 56.769360
std 23.844933
min 10.400000
25% 36.830000
50% 57.940000
75% 74.210000
max 100.000000
desc2 = data['internetuserate'].describe() print (desc2)
count 192.000000
mean 35.632716
std 27.780285
min 0.210066
25% 9.999604
50% 31.810121
75% 56.416046
max 95.638113
#basic scatterplot: Q->Q
scat1 = seaborn.regplot(x="urbanrate", y="internetuserate", fit_reg=False, data=data)
plt.xlabel('Urban Rate')
Text(0.5, 0, 'Urban Rate')
plt.ylabel('Internet Use Rate')
Text(0, 0.5, 'Internet Use Rate')
plt.title('Scatterplot for the Association Between Urban Rate and Internet Use Rate')
Text(0.5, 1.0, 'Scatterplot for the Association Between Urban Rate and Internet Use Rate')
scat2 = seaborn.regplot(x="urbanrate", y="internetuserate", data=data)
#scatterplot graph image cannot be attached here
plt.xlabel('Urban Rate')
Text(0.5, 0, 'Urban Rate')
plt.ylabel('Internet Use Rate')
Text(0, 0.5, 'Internet Use Rate')
plt.title('Scatterplot for the Association Between Urban Rate and Internet Use Rate')
Text(0.5, 1.0, 'Scatterplot for the Association Between Urban Rate and Internet Use Rate')
scat3 = seaborn.regplot(x="incomeperperson", y="internetuserate", data=data)
#scatterplot graph image cannot be attached here
plt.xlabel('Income per Person')
plt.ylabel('Internet Use Rate')
plt.title('Scatterplot for the Association Between Income per Person and Internet Use Rate')
scat4 = seaborn.regplot(x="incomeperperson", y="hivrate", data=data)
plt.xlabel('Income per Person')
plt.ylabel('HIV Rate')
plt.title('Scatterplot for the Association Between Income per Person and HIV Rate')
#quartile split (use qcut function & ask for 4 groups - gives you quartile split)
print ('Income per person - 4 categories - quartiles')
data['INCOMEGRP4']=pandas.qcut(data.incomeperperson, 4, labels=["1=25th%tile","2=50%tile","3=75%tile","4=100%tile"])
c10 = data['INCOMEGRP4'].value_counts(sort=False, dropna=True)
print(c10)
1=25th%tile 48
2=50%tile 47
3=75%tile 47
4=100%tile 48
#bivariate bar graph C->Q
seaborn.catplot(x='INCOMEGRP4', y='hivrate', data=data, kind="bar", ci=None)
#bar chart image cannot be attached here
plt.xlabel('income group')
plt.ylabel('mean HIV rate')
c11= data.groupby('INCOMEGRP4').size()
print (c11)
INCOMEGRP4
1=25th%tile 48
2=50%tile 47
3=75%tile 47
4=100%tile 48
#Summary:
We examined four variables - internateuser rate, urban rate, income per person and HIV rate from gapminder dataset and understand each variables descriptive statistics.
Next we did a co-relation analysis on these variables to find association b/w them. From the urban rate & internet use rate we find there is positive relation b/w them.
plt.xlabel('Urban Rate')
plt.ylabel('Internet Use Rate')
plt.title('Scatterplot for the Association Between Urban Rate and Internet Use Rate')
scat3 = seaborn.regplot(x="incomeperperson", y="internetuserate", data=data)
plt.xlabel('Income per Person')
plt.ylabel('Internet Use Rate')
plt.title('Scatterplot for the Association Between Income per Person and Internet Use Rate')
Text(0.5, 1.0, 'Scatterplot for the Association Between Income per Person and Internet Use Rate')
scat4 = seaborn.regplot(x="incomeperperson", y="hivrate", data=data)
plt.ylabel('HIV Rate')
plt.title('Scatterplot for the Association Between Income per Person and HIV Rate')
0 notes
Text
Assignment 3
import pandas import numpy import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
data = pandas.read_csv('gapminder.csv', low_memory=False)
print (len(data)) #number of observations (rows)
213
#setting variables you will be working with to numeric
data['urbanrate'] = pandas.to_numeric(data['urbanrate'], errors='coerce') data['femaleemployrate'] = pandas.to_numeric(data['femaleemployrate'], errors='coerce') data['lifeexpectancy'] = pandas.to_numeric(data['lifeexpectancy'], errors='coerce')
#creating three new variables for data management operations - urban,fer & le
data['urban']=data['urbanrate'] data['fer']=data['femaleemployrate'] data['le']=data['lifeexpectancy']
#data management for urbanrate
def urban (row): if row['urbanrate'] >= 75: return 3 if 25 <= row['urbanrate'] < 75: return 2 if row['urbanrate'] < 25: return 1
data['urban'] = data.apply(lambda row: urban (row),axis=1)
#frequency distribution for new varaibale urban
c1 = data['urban'].value_counts(sort=False) print (c1)
1.0 22
2.0 133
3.0 48
#data management for femaleemployrate
def fer (row): if row['femaleemployrate'] < 20: return 1 if 20 <= row['femaleemployrate'] < 40: return 2 if 40 <= row['femaleemployrate'] < 60: return 3 if 60 <= row['femaleemployrate'] < 80: return 4 if row['femaleemployrate'] >= 80: return 5
data['fer'] = data.apply(lambda row: fer (row),axis=1)
#frequency distribution for new varaibale fer
c2 = data['fer'].value_counts(sort=False) print (c2)
2.0 45
3.0 96
4.0 26
5.0 4
1.0 7
#data management for le
def le (row): if row['lifeexpectancy'] < 45: return 1 if 45 <= row['lifeexpectancy'] < 55: return 2 if 55 <= row['lifeexpectancy'] < 65: return 3 if 65 <= row['lifeexpectancy'] < 75: return 4 if row['lifeexpectancy'] >= 75: return 5
data['le'] = data.apply(lambda row: le (row),axis=1)
#frequency distribution for new varaibale le
c3 = data['le'].value_counts(sort=False) print (c3)
2.0 24
5.0 65
4.0 76
3.0 26
Summary:
In this module for data management I have collapsed the responses for urbanrate, femaleemployrate, and lifeexpectancy to create three new variables which are: urban, fer, and le.
For urban, the most commonly endorsed response was 2 (62.44%), meaning that most countries have an urban rate of 25% - 75%.
For fer, the most commonly endorsed response was 3 (45.07%), meaning that almost half of the countries have a female employment rate of 40% - 60%.
For le, the most commonly endorsed response was 4 (35.68%), meaning that the life expectancy in about one-third of the countries is 65 – 75 years old.
0 notes
Text
Assignment 2 (week 2)
import pandas import numpy import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
data = pandas.read_csv('gapminder.csv', low_memory=False)
print (len(data)) #number of observations (rows)
213
print (len(data.columns)) # number of variables (columns)
16
setting variables you will be working with to numeric
data['internetuserate'] = pandas.to_numeric(data['internetuserate'], errors='coerce') data['urbanrate'] = pandas.to_numeric(data['urbanrate'], errors='coerce') data['incomeperperson'] = pandas.to_numeric(data['incomeperperson'], errors='coerce')
ADDING TITLES (i.e. frequency distributions) for each variable
print ('counts for internetuserate - 2010 Internet users (per 100 people)') c1 = data['internetuserate'].value_counts(sort=False) print (c1)
counts for internetuserate - 2010 Internet users (per 100 people) 81.000000 1
66.000000 1
45.000000 1
65.000000 1
2.100213 1
..
36.000335 1
41.000128 1
33.616683 1
60.119707 1
1.699985 1
Name: internetuserate, Length: 192, dtype: int64
print ('percentages for internetuserate - 2010 Internet users (per 100 people)') p1 = data['internetuserate'].value_counts(sort=False, normalize=True) print (p1)
percentages for internetuserate - 2010 Internet users (per 100 people)
81.000000 0.005208
66.000000 0.005208
45.000000 0.005208
65.000000 0.005208
2.100213 0.005208
...
36.000335 0.005208
41.000128 0.005208
33.616683 0.005208
60.119707 0.005208
1.699985 0.005208
Name: internetuserate, Length: 192, dtype: float64
print ('counts for urbanrate - 2008 urban poplation (% of total)') c2 = data['urbanrate'].value_counts(sort=False) print(c2)
counts for urbanrate - 2008 urban poplation (% of total)
92.00 1
100.00 6
74.50 1
73.50 1 17.00 1
..
56.02 1
57.18 1
73.92 1
25.46 1
28.38 1
Name: urbanrate, Length: 194, dtype: int64
print ('percentages for urbanrate -2008 urban poplation (% of total)') p2 = data['urbanrate'].value_counts(sort=False, normalize=True) print (p2)
percentages for urbanrate -2008 urban poplation (% of total)
92.00 0.004926
100.00 0.029557
74.50 0.004926
73.50 0.004926
17.00 0.004926
...
56.02 0.004926
57.18 0.004926
73.92 0.004926
25.46 0.004926
28.38 0.004926
Name: urbanrate, Length: 194, dtype: float64
print ('counts for incomeperperson - 2010 GDP per capita in constant 2000 US $') c3 = data['incomeperperson'].value_counts(sort=False, dropna=False) print(c3)
counts for incomeperperson - 2010 GDP per capita in constant 2000 US $
NaN 23 8614.120219 1
39972.352768 1
279.180453 1
161.317137 1
..
377.421113 1
2344.896916 1
25306.187193 1
4180.765821 1
25575.352623 1
Name: incomeperperson, Length: 191, dtype: int64
print ('percentages for incomeperperson - 2010 GDP per capita in constant 2000 US $') p3 = data['incomeperperson'].value_counts(sort=False, normalize=True) print (p3)
percentages for incomeperperson - 2010 GDP per capita in constant 2000 US $
5188.900935 0.005263
8614.120219 0.005263
39972.352768 0.005263
279.180453 0.005263
161.317137 0.005263
...
377.421113 0.005263
2344.896916 0.005263
25306.187193 0.005263
4180.765821 0.005263
25575.352623 0.005263
Name: incomeperperson, Length: 190, dtype: float64
Summary: The data set taken here is 'gapminder.csv'. I have considered three variables of interest here which are:
internetuserate - 2010 Internet users (per 100 people)
urbanrate - 2008 urban poplation (% of total)
incomeperperson - 2010 GDP per capita in constant 2000 US $
The program is written and compiled in python and output snapshot is provided above. First I have run the # of observations in the data set (rows) which is 213. Then have run the # of variables in the data set (columns) whose value is 16.
Next, as per the assignment requirement, I have run the frequency distribution (i.e counts and percentages of all these three variables considered) and provided the snapshot.
0 notes
Text
Assignment 1 - Data Management & Visualization
Data Set Selected: I would like to work on the Gapminder data set which contains data for all 192 UN members and which seeks to increase the use and understanding of statistics about social, economic, and environmental development at local, national, and global levels.
2. Research Question: Determine associations (if any) between "rate of internet users" by the "rate of the country's population living in urban settings".
Variables of interest from the codebook for the above stated hypothesis - Internetuserate & Urbanrate
Variables Type - Both are ordinal variables
3. Second hypothesis: Is there any association b/w rate of internet users and income per person for a region/country?
Variables of interest from the codebook for the above stated hypothesis - Internetuserate & Incomeperperson
Variables Type - Both are ordinal variables
4. Literature review of hypothesis #1: IJERPH | Free Full-Text | Association between Urban Upbringing and Compulsive Internet Use in Japan: A Cross-Sectional, Multilevel Study with Retrospective Recall (mdpi.com)
5. From the above literature review, the findings as are below:
Growing up in a large city was significantly associated with higher Compulsive Internet Use Scale (CIUS) scores (γ = 1.65, Standard Error (SE) = 0.45) and Mild CIU + Severe CIU (Exp(γ) = 1.44; 95% Confidence Interval (CI) (1.04–2.00)) compared to growing up in a small municipality
0 notes
Text
Statistical Interactions - Assignment4
Objective:
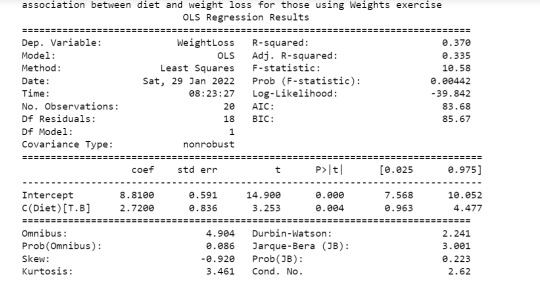
The goal of this assignment is to perform data analysis on a dataset ("diet_exercise.csv") to reveal any association that may exist b/w weight-loss and diet. Here the categorical variable is "Diet" given by two levels - Diet A & Diet B. The quantitative variable is weigh-tloss. The study is performed on these two variables using ANOVA method and results show below (first part) the p value is 0.00133 which is less than 0.05 which means there is slight significance b/w these two variables.
Now as part of this exercise we also wanted to introduce a moderator variable called "exercise" which has two levels - Cardio & Weights.
We ran the same tests with the introduction of the third moderator variable called "exercise" and found the below results.
Results:
The weight loss was shown to be significant for subjects who were on cardio exercise (p value of 2.00e-12) compared to those who used weights (p value of 0.00442). Hence the moderator variable is useful to show the actual relationship b/w two variables may be due to a 3rd moderator variable.
Code Snippet & Output:
import numpy
import pandas
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
import seaborn
import matplotlib.pyplot as plt
data = pandas.read_csv('diet_exercise.csv', low_memory=False)
model1 = smf.ols(formula='WeightLoss ~ C(Diet)', data=data).fit()
print (model1.summary())
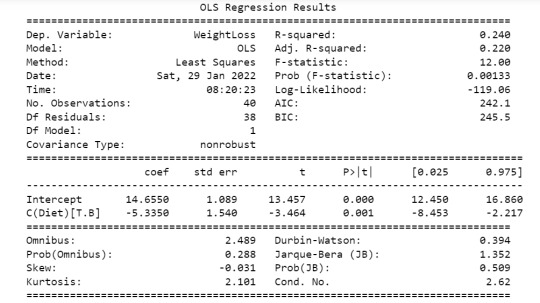
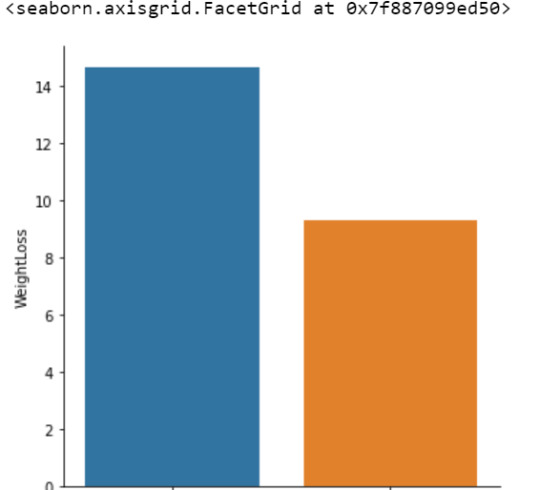
sub1 = data[['WeightLoss', 'Diet']].dropna()
m1= sub1.groupby('Diet').mean()
print ("means for WeightLoss by Diet A vs. B")
print (m1)
means for WeightLoss by Diet A vs. B
WeightLoss
Diet
A 14.655
B 9.320
st1= sub1.groupby('Diet').std()
print ("standard deviation for mean WeightLoss by Diet A vs. B")
print (st1)
standard deviation for mean WeightLoss by Diet A vs. B
WeightLoss
Diet
A 6.302086
B 2.779360
# bivariate bar graph
seaborn.factorplot(x="Diet", y="WeightLoss", data=data, kind="bar", ci=None)
plt.xlabel('Diet Type')
plt.ylabel('Mean Weight Loss in pounds')
sub2=data[(data['Exercise']=='Cardio')]
sub3=data[(data['Exercise']=='Weights')]
model2 = smf.ols(formula='WeightLoss ~ C(Diet)', data=sub2).fit()
print ('association between diet and weight loss for those using Cardio exercise')
print (model2.summary())

model3 = smf.ols(formula='WeightLoss ~ C(Diet)', data=sub3).fit()
print ('association between diet and weight loss for those using Weights exercise')
print (model3.summary())
m3= sub2.groupby('Diet').mean()
print ("means for WeightLoss by Diet A vs. B for CARDIO")
print (m3)

m4 = sub3.groupby('Diet').mean()
print ("Means for WeightLoss by Diet A vs. B for WEIGHTS")
print (m4)

0 notes
Text
Assignment 3 - Calculating a Correlation Co-efficient (r):
Objective:
For this assignment we needed to generate a correlation coefficient from our data among the variables of interest. In this study we were interested to see if there was any association between 'Urban rate' & 'Internet use rate' as well as 'Income per person' & 'Internet use rate'. All three data sets are quantitative. Python was used to conduct the analysis.
Results:
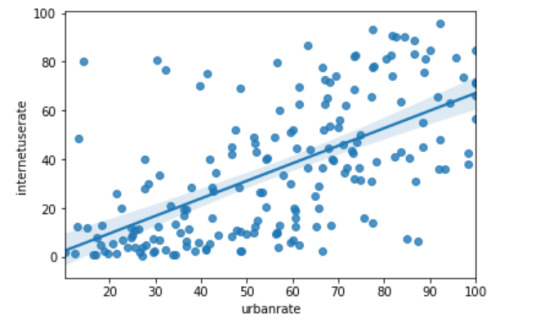
1) Urban rate & internet use rate: r (Correlation coefficient) = 0.62 and P value = 4.540316299446745e-21. Since the r value is close to 1 and the p value is very small, this tells us that the relationship is statistically significant between urbanization rate and internet use rate.
2) Income per person & internet use rate: r (Correlation coefficient) = 0.75 and P value = 3.067338031233027e-34. Since the r value is close to 1 and the p value is very small, this tells us that the relationship is statistically significant between income per person and internet use rate.
Code Snippet:
import pandas
import numpy
import seaborn
import scipy
import matplotlib.pyplot as plt
data = pandas.read_csv('gapminder.csv', low_memory=False)
#setting variables you will be working with to numeric
data['internetuserate'] = pandas.to_numeric(data['internetuserate'], errors='coerce')
data['urbanrate'] = pandas.to_numeric(data['urbanrate'], errors='coerce')
data['incomeperperson'] = pandas.to_numeric(data['incomeperperson'], errors='coerce')
data['incomeperperson']=data['incomeperperson'].replace(' ', numpy.nan)
scat1 = seaborn.regplot(x="urbanrate", y="internetuserate", fit_reg=True, data=data)
plt.xlabel('Urban Rate')
plt.ylabel('Internet Use Rate')
plt.title('Scatterplot for the Association Between Urban Rate and Internet Use Rate')
scat2 = seaborn.regplot(x="incomeperperson", y="internetuserate", fit_reg=True, data=data)
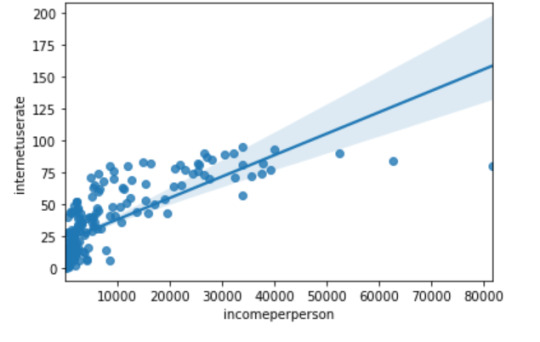
plt.xlabel('Income per Person')
plt.ylabel('Internet Use Rate')
plt.title('Scatterplot for the Association Between Income per Person and Internet Use Rate')
data_clean=data.dropna()
print ('association between urbanrate and internetuserate')
print (scipy.stats.pearsonr(data_clean['urbanrate'], data_clean['internetuserate']))
association between urbanrate and internetuserate
(0.6244640029489794, 4.540316299446745e-21)
print ('association between incomeperperson and internetuserate')
print (scipy.stats.pearsonr(data_clean['incomeperperson'], data_clean['internetuserate']))
association between incomeperperson and internetuserate (0.7507274333051273, 3.067338031233027e-34)
0 notes
Text
Assignment 2
Objective:
For this assignment a Chi-square Test of Independence in Python was supposed to be executed to analyze association b/w two categorical variables - 'Nicotine Dependence' & 'Days smoked per month'. The dataset "NESACR.csv" was used to see if there is an association b/w nicotine dependence proportion & cigarettes smoked per month for young adults in age bracket of 18 to 25. In other words if there is an increase in nicotine dependence when the # of cigarettes smoked in a month increased. There were six levels for the cigarettes smoked per month - 1, 2.5, 6, 14, 22 & 30. We did run a post-hoc analysis b/w each of the pairs i.e nicotine dependence within each smoking frequency group.
Results:
The results shows the p value is significant when the # cigarettes smoked increased. The chi-square value, p value & expected counts for each smoking frequency group is shown below.
Code Snippet & Output:
import pandas
import numpy
import scipy.stats
import seaborn
import matplotlib.pyplot as plt
data = pandas.read_csv('nesarc.csv', low_memory=False)
# new code setting variables you will be working with to numeric
data['TAB12MDX'] = pandas.to_numeric(data['TAB12MDX'], errors='coerce')
data['CHECK321'] = pandas.to_numeric(data['CHECK321'], errors='coerce')
data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1'], errors='coerce')
data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1'], errors='coerce')
data['AGE'] = pandas.to_numeric(data['AGE'], errors='coerce')
#subset data to young adults age 18 to 25 who have smoked in the past 12 months
sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
#make a copy of my new subsetted data
sub2 = sub1.copy()
# recode missing values to python missing (NaN)
sub2['S3AQ3B1']=sub2['S3AQ3B1'].replace(9, numpy.nan)
sub2['S3AQ3C1']=sub2['S3AQ3C1'].replace(99, numpy.nan)
#recoding values for S3AQ3B1 into a new variable, USFREQMO
recode1 = {1: 30, 2: 22, 3: 14, 4: 6, 5: 2.5, 6: 1}
sub2['USFREQMO']= sub2['S3AQ3B1'].map(recode1)
# contingency table of observed counts
ct1=pandas.crosstab(sub2['TAB12MDX'], sub2['USFREQMO'])
print (ct1)
USFREQMO 1.0 2.5 6.0 14.0 22.0 30.0
TAB12MDX
0 64 53 69 59 41 521
1 7 12 19 32 27 799
# column percentages
colsum=ct1.sum(axis=0)
colpct=ct1/colsum
print(colpct)
USFREQMO 1.0 2.5 6.0 14.0 22.0 30.0
TAB12MDX
0 0.901408 0.815385 0.784091 0.648352 0.602941 0.394697
1 0.098592 0.184615 0.215909 0.351648 0.397059 0.605303
# chi-square
print ('chi-square value, p value, expected counts')
cs1= scipy.stats.chi2_contingency(ct1)
print (cs1)
chi-square value, p value, expected counts
(165.27320708055845, 7.436364208390599e-34, 5, array([[ 33.64474457, 30.80152672, 41.70052848, 43.1221374 , 32.22313564, 625.50792719], [ 37.35525543, 34.19847328, 46.29947152, 47.8778626 , 35.77686436, 694.49207281]]))
# set variable types
sub2["USFREQMO"] = sub2["USFREQMO"].astype('category')
sub2['TAB12MDX'] = pandas.to_numeric(sub2['TAB12MDX'], errors='coerce')
# graph percent with nicotine dependence within each smoking frequency group
seaborn.factorplot(x="USFREQMO", y="TAB12MDX", data=sub2, kind="bar", ci=None)
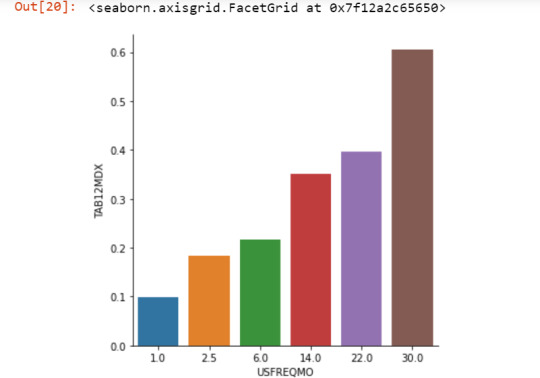
plt.xlabel('Days smoked per month')
plt.ylabel('Proportion Nicotine Dependent')
recode2 = {1: 1, 2.5: 2.5}
sub2['COMP1v2']= sub2['USFREQMO'].map(recode2)
ct2=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP1v2'])
print (ct2)
COMP1v2 1.0 2.5
TAB12MDX
0 64 53
1 7 12
# column percentages
colsum=ct2.sum(axis=0)
colpct=ct2/colsum
print(colpct)
COMP1v2 1.0 2.5
TAB12MDX
0 0.901408 0.815385
1 0.098592 0.184615
print ('chi-square value, p value, expected counts')
cs2= scipy.stats.chi2_contingency(ct2)
print (cs2)
chi-square value, p value, expected counts
(1.4348930637007287, 0.2309675448977717, 1, array([[61.08088235, 55.91911765], [ 9.91911765, 9.08088235]]))
recode3 = {1: 1, 6: 6}
sub2['COMP1v6']= sub2['USFREQMO'].map(recode3)
# contingency table of observed counts
ct3=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP1v6'])
print (ct3)
COMP1v6 1.0 6.0
TAB12MDX
0 64 69
1 7 19
# column percentages
colsum=ct3.sum(axis=0)
colpct=ct3/colsum
print(colpct)
COMP1v6 1.0 6.0
TAB12MDX
0 0.901408 0.784091
1 0.098592 0.215909
print ('chi-square value, p value, expected counts')
cs3= scipy.stats.chi2_contingency(ct3)
print (cs3)
chi-square value, p value, expected counts
(3.142840191220936, 0.07626090198286821, 1, array([[59.38993711, 73.61006289], [11.61006289, 14.38993711]]))
recode4 = {1: 1, 14: 14}
sub2['COMP1v14']= sub2['USFREQMO'].map(recode4)
# contingency table of observed counts
ct4=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP1v14'])
print (ct4)
COMP1v14 1.0 14.0
TAB12MDX
0 64 59
1 7 32
# column percentages
colsum=ct4.sum(axis=0)
colpct=ct4/colsum
print(colpct)
COMP1v14 1.0 14.0
TAB12MDX
0 0.901408 0.648352
1 0.098592 0.351648
print ('chi-square value, p value, expected counts')
cs4= scipy.stats.chi2_contingency(ct4)
print (cs4)
chi-square value, p value, expected counts
(12.622564075461572, 0.00038111819882681824, 1, array([[53.90740741, 69.09259259], [17.09259259, 21.90740741]]))
recode5 = {1: 1, 22: 22}
sub2['COMP1v22']= sub2['USFREQMO'].map(recode5)
# contingency table of observed counts
ct5=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP1v22'])
print (ct5)
COMP1v22 1.0 22.0
TAB12MDX
0 64 41
1 7 27
# column percentages
colsum=ct5.sum(axis=0)
colpct=ct5/colsum
print(colpct)
COMP1v22 1.0 22.0
TAB12MDX
0 0.901408 0.602941
1 0.098592 0.397059
print ('chi-square value, p value, expected counts')
cs5= scipy.stats.chi2_contingency(ct5)
print (cs5)
chi-square value, p value, expected counts
(15.169488833230059, 9.827865291318501e-05, 1, array([[53.63309353, 51.36690647], [17.36690647, 16.63309353]]))
recode6 = {1: 1, 30: 30}
sub2['COMP1v30']= sub2['USFREQMO'].map(recode6)
# contingency table of observed counts
ct6=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP1v30'])
print (ct6)
COMP1v30 1.0 30.0
TAB12MDX
0 64 521
1 7 799
# column percentages
colsum=ct6.sum(axis=0)
colpct=ct6/colsum
print(colpct)
COMP1v30 1.0 30.0
TAB12MDX
0 0.901408 0.394697
1 0.098592 0.605303
print ('chi-square value, p value, expected counts')
cs6= scipy.stats.chi2_contingency(ct6)
print (cs6)
chi-square value, p value, expected counts
(68.92471874488487, 1.0229460827061155e-16, 1, array([[ 29.85981308, 555.14018692], [ 41.14018692, 764.85981308]]))
recode7 = {2.5: 2.5, 6: 6}
sub2['COMP2v6']= sub2['USFREQMO'].map(recode7)
# contingency table of observed counts
ct7=pandas.crosstab(sub2['TAB12MDX'], sub2['COMP2v6'])
print (ct7)
COMP2v6 2.5 6.0
TAB12MDX
0 53 69
1 12 19
# column percentages
colsum=ct7.sum(axis=0)
colpct=ct7/colsum
print(colpct)
COMP2v6 2.5 6.0
TAB12MDX
0 0.815385 0.784091
1 0.184615 0.215909
print ('chi-square value, p value, expected counts')
cs7=scipy.stats.chi2_contingency(ct7)
print (cs7)
chi-square value, p value, expected counts (0.07430561076945266, 0.7851679729700605, 1, array([[51.83006536, 70.16993464], [13.16993464, 17.83006536]]))
0 notes
Text
Assignment 1
import numpy import pandas import statsmodels.formula.api as smf import statsmodels.stats.multicomp as multi
data = pandas.read_csv('nesarc.csv', low_memory=False)
#setting variables you will be working with to numeric data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1'], errors='coerce') data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1'], errors='coerce') data['CHECK321'] = pandas.to_numeric(data['CHECK321'], errors='coerce')
#subset data to young adults age 18 to 25 who have smoked in the past 12 months sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
#SETTING MISSING DATA sub1['S3AQ3B1']=sub1['S3AQ3B1'].replace(9, numpy.nan) sub1['S3AQ3C1']=sub1['S3AQ3C1'].replace(99, numpy.nan)
#recoding number of days smoked in the past month recode1 = {1: 30, 2: 22, 3: 14, 4: 5, 5: 2.5, 6: 1} sub1['USFREQMO']= sub1['S3AQ3B1'].map(recode1)
#converting new variable USFREQMMO to numeric sub1['USFREQMO'] = pandas.to_numeric(sub1['USFREQMO'], errors='coerce')
# Creating a secondary variable multiplying the days smoked/month and the number of cig/per day sub1['NUMCIGMO_EST']=sub1['USFREQMO'] * sub1['S3AQ3C1'] sub1['NUMCIGMO_EST']= pandas.to_numeric(sub1['NUMCIGMO_EST'], errors='coerce')
ct1 = sub1.groupby('NUMCIGMO_EST').size() print (ct1)
NUMCIGMO_EST
1.0 29
2.0 14
2.5 11
3.0 12
4.0 2
..
1050.0 1
1200.0 29
1800.0 2
2400.0 1
2940.0 1
Length: 66, dtype: int64
# using ols function for calculating the F-statistic and associated p value model1 = smf.ols(formula='NUMCIGMO_EST ~ C(MAJORDEPLIFE)', data=sub1) results1 = model1.fit() print (results1.summary())
OLS Regression Results ================================================
Dep. Variable: NUMCIGMO_EST R-squared: 0.002
Model: OLS Adj. R-squared: 0.002
Method: Least Squares F-statistic: 3.550
Date: Wed, 26 Jan 2022 Prob (F-statistic): 0.0597
Time: 17:58:07 Log-Likelihood: -11934.
No. Observations: 1697 AIC: 2.387e+04
Df Residuals: 1695 BIC: 2.388e+04
Df Model: 1
Covariance Type: nonrobust ================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 312.8380 7.747 40.381 0.000 297.643 328.033
C(MAJORDEPLIFE)[T.1] 28.5370 15.146 1.884 0.060 -1.169 58.243 ================================================
Omnibus: 673.875 Durbin-Watson: 1.982
Prob(Omnibus): 0.000 Jarque-Bera (JB): 5043.141
Skew: 1.672 Prob(JB): 0.00
Kurtosis: 10.755 Cond. No. 2.46 ================================================
Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
sub2 = sub1[['NUMCIGMO_EST', 'MAJORDEPLIFE']].dropna()
print ('means for numcigmo_est by major depression status') m1= sub2.groupby('MAJORDEPLIFE').mean() print (m1)
means for numcigmo_est by major depression status
NUMCIGMO_EST
MAJORDEPLIFE
0 312.837989
1 341.375000
print ('standard deviations for numcigmo_est by major depression status') sd1 = sub2.groupby('MAJORDEPLIFE').std() print (sd1)
standard deviations for numcigmo_est by major depression status NUMCIGMO_EST
MAJORDEPLIFE
0 269.002344
1 288.495118 sub3 = sub1[['NUMCIGMO_EST', 'ETHRACE2A']].dropna()
model2 = smf.ols(formula='NUMCIGMO_EST ~ C(ETHRACE2A)', data=sub3).fit() print (model2.summary())
OLS Regression Results ================================================
Dep. Variable: NUMCIGMO_EST R-squared: 0.055
Model: OLS Adj. R-squared: 0.052
Method: Least Squares F-statistic: 24.40
Date: Wed, 26 Jan 2022 Prob (F-statistic): 1.18e-19
Time: 18:01:14 Log-Likelihood: -11888.
No. Observations: 1697 AIC: 2.379e+04
Df Residuals: 1692 BIC: 2.381e+04
Df Model: 4
Covariance Type: nonrobust ============================================================
coef std err t P>|t| [0.025 0.975]
-------------------------------------------------------------------------------------
Intercept 368.7865 8.229 44.814 0.000 352.646 384.927 C(ETHRACE2A)[T.2] -109.5127 20.189 -5.424 0.000 -149.111 -69.914 C(ETHRACE2A)[T.3] -57.7984 42.038 -1.375 0.169 -140.250 24.653 C(ETHRACE2A)[T.4] -124.5279 36.033 -3.456 0.001 -195.201 -53.854 C(ETHRACE2A)[T.5] -149.0283 16.795 -8.873 0.000 -181.969 -116.087 ============================================================ Omnibus: 712.397 Durbin-Watson: 1.994
Prob(Omnibus): 0.000 Jarque-Bera (JB): 6548.614
Skew: 1.717 Prob(JB): 0.00
Kurtosis: 11.990 Cond. No. 6.72 ============================================================Warnings: [1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
print ('means for numcigmo_est by major depression status') m2= sub3.groupby('ETHRACE2A').mean() print (m2)
means for numcigmo_est by major depression status
NUMCIGMO_EST
ETHRACE2A
1 368.786528
2 259.273810
3 310.988095
4 244.258621
5 219.758258
print ('standard deviations for numcigmo_est by major depression status') sd2 = sub3.groupby('ETHRACE2A').std() print (sd2)
standard deviations for numcigmo_est by major depression status NUMCIGMO_EST
ETHRACE2A
1 281.430730
2 278.677392
3 260.116964
4 195.076441
5 220.859365
mc1 = multi.MultiComparison(sub3['NUMCIGMO_EST'], sub3['ETHRACE2A']) res1 = mc1.tukeyhsd() print(res1.summary())
Multiple Comparison of Means - Tukey HSD, FWER=0.05 ========================================================= group1 group2 meandiff p-adj lower upper reject
----- ---------------------------------------------------------
1 2 -109.5127 0.001 -164.6441 -54.3814 True
1 3 -57.7984 0.6251 -172.5914 56.9945 False
1 4 -124.5279 0.0051 -222.9229 -26.1329 True
1 5 -149.0283 0.001 -194.89 -103.1665 True
2 3 51.7143 0.7555 -71.6021 175.0307 False
2 4 -15.0152 0.9 -123.233 93.2026 False
2 5 -39.5156 0.4492 -103.8025 24.7714 False
3 4 -66.7295 0.7058 -214.5437 81.0848 False
3 5 -91.2298 0.2269 -210.6902 28.2305 False
4 5 -24.5004 0.9 -128.3027 79.302 False
---------------------------------------------------------
1 note
·
View note