#43093
Quote
少年アシベのネタで「オシャレなレストランを開くことが夢だったけど、別に料理が好きなわけではないので、店で出してる料理は全部レトルトと冷凍食品だし、ケーキは別の店で買ったのそのまま出してる。あくまでオシャレな店をやりたい」っていうのあったけど、ああいう人割といるんじゃないかと思う。
Xユーザーの織部ゆたかさん
27 notes
·
View notes
Text
Running a Lasso Regression Analysis to identify a subset of variables that best predicted the alcohol drinking behavior of individuals
A lasso regression analysis is performed to detect a subgroup of variables from a set of 23 categorical and quantitative predictor variables in the Nesarc Wave 1 dataset which best predicted a quantitative response variable assessing the alcohol drinking behaviour of individuals 18 years and older in terms of number of any alcohol drunk per month.
Categorical predictors include a series of 5 binary categorical variables for ethnicity (Hispanic, White, Black, Native American and Asian) to improve interpretability of the selected model with fewer predictors.
Other binary categorical predictors are related to substance consumption, and in particular to the fact whether or not individuals are lifetime opioids, cannabis, cocaine, sedatives, tranquilizers consumers, as well as suffer from depression.
About dependencies, other binary categorical variables are those regarding if individuals
are lifetime affected by nicotine dependency,
abuse lifetime of alcohol
or not.
Quantitative predictor variables include age when the aforementioned substances have been taken for the first time, the usual frequency of cigarettes and of any alcohol drunk per month, the number of cigarettes smoked per month.
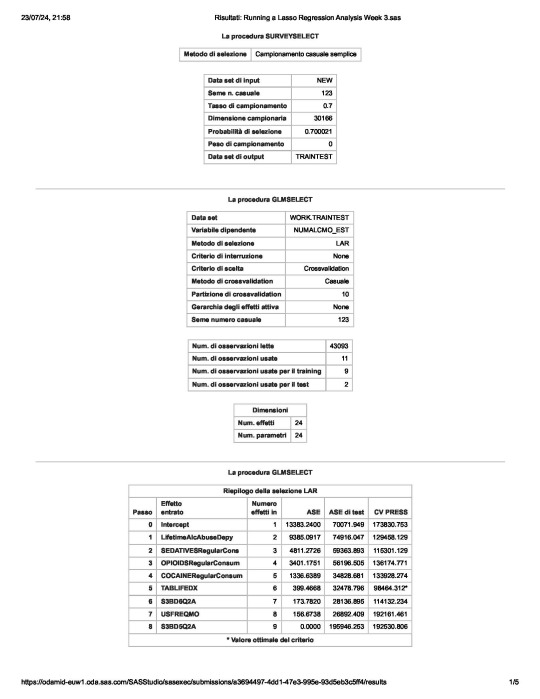
In a nutshell, looking at the SAS output,
the survey select procedure, used to split the observations in the dataset into training and test data, shows that the sample size is 30,166.
the so-called “NUMALCMO_EST” dependent variable - number of any alcohol drunk per month - along with the selection method used, are being displayed, together with information such as
the choice of a cross validation criteria as criterion for choosing the best model, with K equals 10-fold cross validation,
the random assignments of observations to the folds
the total number of observations in the data set are 43093 and the number of observations used for training and testing the statistical models are 11, where 9 are for training and 2 for testing
the number of parameters to be estimated is 24 for the intercept plus the 23 predictors.
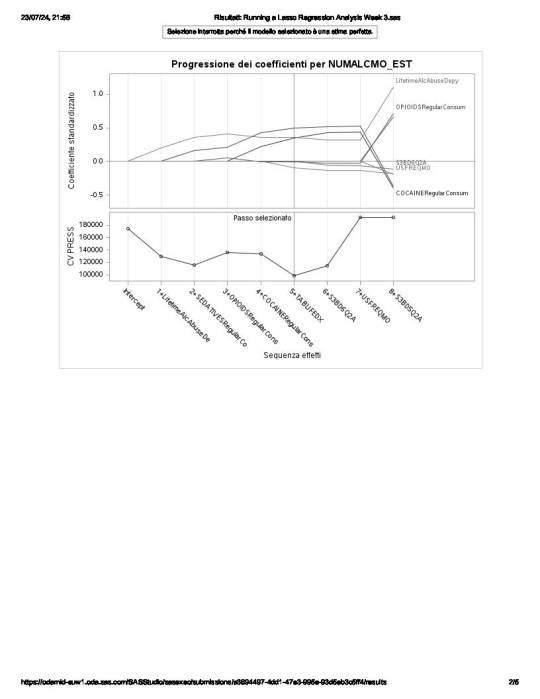
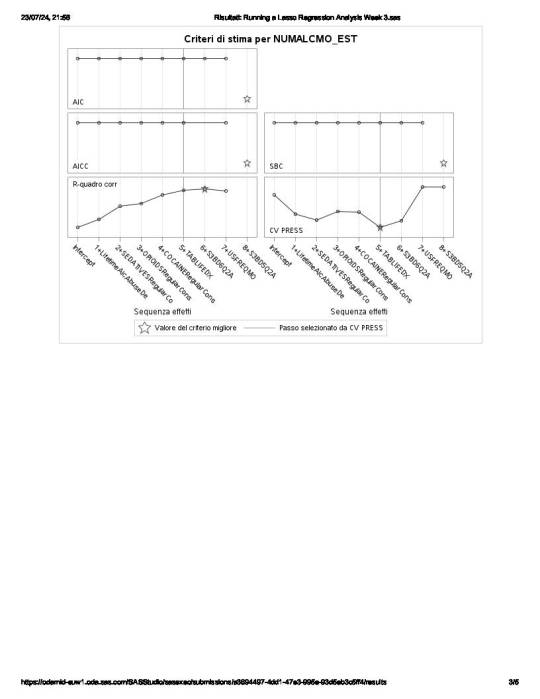
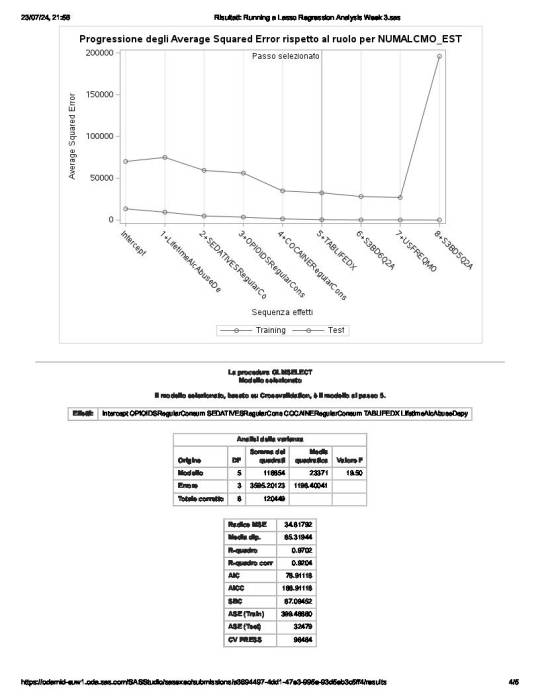
of the 23 predictor variables, 8 have been maintained in the selected model:
LifetimeAlcAbuseDepy - alcohol abuse / dependence both in last 12 months and prior to the last 12 months
and
OPIOIDSRegularConsumer – used opioids both in the last 12 months and prior to the last 12 months
have the largest regression coefficient, followed by COCAINERegularConsum behaviour.
LifetimeAlcAbuseDepy and OPIOIDSRegularConsum are positively associated with the response variable, while COCAINERegularConsum is negatively associated with NUMALCMO_EST.
Other predictors associated with lower number of any alcohol drunk per month are S3BD6Q2A – “age first used cocaine or crack” and USEFREQMO – “usual frequency of cigarettes smoked per month”.
These 8 variables accounted for 33.6% of the variance in the number of any alcohol drunk per month response variable.
Hereunder the SAS code used to generate the present analysis and the plots on which the above described results are depicted
PROC IMPORT DATAFILE ='/home/u63783903/my_courses/nesarc_pds.csv' OUT = imported REPLACE;
RUN;
DATA new;
set imported;
/* lib name statement and data step to call in the
NESARC data set for the purpose of growing decision trees*/
LABEL MAJORDEPLIFE = "MAJOR DEPRESSION - LIFETIME"
ETHRACE2A = "IMPUTED RACE/ETHNICITY"
WhiteGroup = "White, Not Hispanic or Latino"
BlackGroup = "Black, Not Hispanic or Latino"
NamericaGroup = "American Indian/Alaska Native, Not Hispanic or Latino"
AsianGroup = "Asian/Native Hawaiian/Pacific Islander, Not Hispanic or Latino"
HispanicGroup = "Hispanic or Latino"
S3BD3Q2B = "USED OPIOIDS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS"
OPIOIDSRegularConsumer = "USED OPIOIDS both IN THE LAST 12 MONTHS AND PRIOR TO LAST 12 MONTHS"
S3BD3Q2A = "AGE FIRST USED OPIOIDS"
S3BD5Q2B = "USED CANNABIS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS"
CANNABISRegularConsumer = "USED CANNABIS both IN THE LAST 12 MONTHS AND PRIOR TO LAST 12 MONTHS"
S3BD5Q2A = "AGE FIRST USED CANNABIS"
S3BD1Q2B = "USED SEDATIVES IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS"
SEDATIVESRegularConsumer = "USED SEDATIVES both IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS"
S3BD1Q2A = "AGE FIRST USED SEDATIVES"
S3BD2Q2B = "USED TRANQUILIZERS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS"
TRANQUILIZERSRegularConsumer = "USED TRANQUILIZERS both IN THE LAST 12 MONTHS AND PRIOR TO LAST 12 MONTHS"
S3BD2Q2A = "AGE FIRST USED TRANQUILIZERS"
S3BD6Q2B = "USED COCAINE OR CRACK IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS"
COCAINERegularConsumer = "USED COCAINE both IN THE LAST 12 MONTHS AND PRIOR TO LAST 12 MONTHS"
S3BD6Q2A = "AGE FIRST USED COCAINE OR CRACK"
TABLIFEDX = "NICOTINE DEPENDENCE - LIFETIME"
ALCABDEP12DX = "ALCOHOL ABUSE/DEPENDENCE IN LAST 12 MONTHS"
ALCABDEPP12DX = "ALCOHOL ABUSE/DEPENDENCE PRIOR TO THE LAST 12 MONTHS"
LifetimeAlcAbuseDepy = "ALCOHOL ABUSE/DEPENDENCE both IN LAST 12 MONTHS and PRIOR TO THE LAST 12 MONTHS"
S3AQ3C1 = "USUAL QUANTITY WHEN SMOKED CIGARETTES"
S3AQ3B1 = "USUAL FREQUENCY WHEN SMOKED CIGARETTES"
USFREQMO = "usual frequency of cigarettes smoked per month"
NUMCIGMO_EST= "NUMBER OF cigarettes smoked per month"
S2AQ8A = "HOW OFTEN DRANK ANY ALCOHOL IN LAST 12 MONTHS"
S2AQ8B = "NUMBER OF DRINKS OF ANY ALCOHOL USUALLY CONSUMED ON DAYS WHEN DRANK ALCOHOL IN LAST 12 MONTHS"
USFREQALCMO = "usual frequency of any alcohol drunk per month"
NUMALCMO_EST = "NUMBER OF ANY ALCOHOL drunk per month"
S3BD1Q2E = "HOW OFTEN USED SEDATIVES WHEN USING THE MOST"
USFREQSEDATIVESMO = "usual frequency of any alcohol drunk per month"
S1Q1F = "BORN IN UNITED STATES"
if cmiss(of _all_) then delete;
/* delete observations with missing data on any of the variables in the NESARC dataset */
if ETHRACE2A=1 then WhiteGroup=1;
else WhiteGroup=0;
/* creation of a variable for white ethnicity coded for 0 for non white ethnicity and 1 for white ethnicity */
if ETHRACE2A=2 then BlackGroup=1;
else BlackGroup=0;
/* creation of a variable for black ethnicity coded for 0 for non black ethnicity and 1 for black ethnicity */
if ETHRACE2A=3 then NamericaGroup=1;
else NamericaGrouGroup=0;
/* same for native american ethnicity*/
if ETHRACE2A=4 then AsianGroup=1;
else AsianGroup=0;
/* same for asian ethnicity */
if ETHRACE2A=5 then HispanicGroup=1;
else HispanicGroup=0;
/* same for hispanic ethnicity */
if S3BD3Q2B = 9 then S3BD3Q2B = .;
/* unknown observations set to missing data wrt usage of OPIOIDS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS */
if S3BD3Q2B = 3 then OPIOIDSRegularConsumer = 1;
if S3BD3Q2B = 1 or S3BD3Q2B = 2 then OPIOIDSRegularConsumer = 0;
if S3BD3Q2B = . then OPIOIDSRegularConsumer = .;
/* creation of a group variable where lifetime opioids consumers are coded to 1
and 0 for non lifetime opioids consumers */
if S3BD3Q2A = 99 then S3BD3Q2A = . ;
/* unknown observations set to missing data wrt AGE FIRST USED OPIOIDS */
if S3BD5Q2B = 9 then S3BD5Q2B = .;
/* unknown observations set to missing data wrt usage of CANNABIS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS */
if S3BD5Q2B = 3 then CANNABISRegularConsumer = 1;
if S3BD5Q2B = 1 or S3BD5Q2B = 2 then CANNABISRegularConsumer = 0;
if S3BD5Q2B = . then CANNABISRegularConsumer = .;
/* creation of a group variable where lifetime cannabis consumers are coded to 1
and 0 for non lifetime cannabis consumers */
if S3BD5Q2A = 99 then S3BD5Q2A = . ;
/* unknown observations set to missing data wrt AGE FIRST USED CANNABIS */
if S3BD1Q2B = 9 then S3BD1Q2B = .;
/* unknown observations set to missing data wrt usage of SEDATIVES IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS */
if S3BD1Q2B = 3 then SEDATIVESRegularConsumer = 1;
if S3BD1Q2B = 1 or S3BD1Q2B = 2 then SEDATIVESRegularConsumer = 0;
if S3BD1Q2B = . then SEDATIVESRegularConsumer = .;
/* creation of a group variable where lifetime sedatives consumers are coded to 1
and 0 for non lifetime sedatives consumers */
if S3BD1Q2A = 99 then S3BD1Q2A = . ;
/* unknown observations set to missing data wrt AGE FIRST USED sedatives */
if S3BD2Q2B = 9 then S3BD1Q2B = .;
/* unknown observations set to missing data wrt usage of TRANQUILIZERS IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS */
if S3BD2Q2B = 3 then TRANQUILIZERSRegularConsumer = 1;
if S3BD2Q2B = 1 or S3BD2Q2B = 2 then TRANQUILIZERSRegularConsumer = 0;
if S3BD2Q2B = . then TRANQUILIZERSRegularConsumer = .;
/* creation of a group variable where lifetime TRANQUILIZERS consumers are coded to 1
and 0 for non lifetime TRANQUILIZERS consumers */
if S3BD2Q2A = 99 then S3BD2Q2A = . ;
/* unknown observations set to missing data wrt AGE FIRST USED TRANQUILIZERS */
if S3BD6Q2A = 9 then S3BD6Q2A = .;
/* unknown observations set to missing data wrt usage of COCAINE IN THE LAST 12 MONTHS/PRIOR TO LAST 12 MONTHS/BOTH TIME PERIODS */
if S3BD6Q2B = 3 then COCAINERegularConsumer = 1;
if S3BD6Q2B = 1 or S3BD6Q2B = 2 then COCAINERegularConsumer = 0;
if S3BD6Q2B = . then COCAINERegularConsumer = .;
/* creation of a group variable where lifetime COCAINE consumers are coded to 1
and 0 for non lifetime COCAINE consumers */
if S3BD6Q2A = 99 then S3BD2Q2A = . ;
/* unknown observations set to missing data wrt AGE FIRST USED COCAINE */
if ALCABDEP12DX = 3 and ALCABDEPP12DX = 3 then LifetimeAlcAbuseDepy =1;
else LifetimeAlcAbuseDepy = 0;
/* creation of a group variable where consumers with lifetime alcohol abuse and dependence are coded to 1
and 0 for consumers with no lifetime alcohol abuse and dependence */
if S3AQ3C1=99 THEN S3AQ3C1=.;
IF S3AQ3B1=9 THEN SS3AQ3B1=.;
IF S3AQ3B1=1 THEN USFREQMO=30;
ELSE IF S3AQ3B1=2 THEN USFREQMO=22;
ELSE IF S3AQ3B1=3 THEN USFREQMO=14;
ELSE IF S3AQ3B1=4 THEN USFREQMO=5;
ELSE IF S3AQ3B1=5 THEN USFREQMO=2.5;
ELSE IF S3AQ3B1=6 THEN USFREQMO=1;
/* usual frequency of smoking per month */
NUMCIGMO_EST=USFREQMO*S3AQ3C1;
/* number of cigarettes smoked per month */
if S2AQ8A=99 THEN S2AQ8A=.;
if S2AQ8B = 99 then S2AQ8B = . ;
IF S2AQ8A=1 THEN USFREQALCMO=30;
ELSE IF S2AQ8A=2 THEN USFREQALCMO=30;
ELSE IF S2AQ8A=3 THEN USFREQALCMO=14;
ELSE IF S2AQ8A=4 THEN USFREQALCMO=8;
ELSE IF S2AQ8A=5 THEN USFREQALCMO=4;
ELSE IF S2AQ8A=6 THEN USFREQALCMO=2.5;
ELSE IF S2AQ8A=7 THEN USFREQALCMO=1;
ELSE IF S2AQ8A=8 THEN USFREQALCMO=0.75;
ELSE IF S2AQ8A=9 THEN USFREQALCMO=0.375;
ELSE IF S2AQ8A=10 THEN USFREQALCMO=0.125;
/* usual frquency of alcohol drinking per month */
NUMALCMO_EST=USFREQALCMO*S2AQ8B;
/* number of any alcohol drunk per month */
if S3BD1Q2E=99 THEN S3BD1Q2E=.;
IF S3BD1Q2E=1 THEN USFREQSEDATIVESMO=30;
ELSE IF S3BD1Q2E=2 THEN USFREQSEDATIVESMO=30;
ELSE IF S3BD1Q2E=3 THEN USFREQSEDATIVESMO=14;
ELSE IF S3BD1Q2E=4 THEN USFREQSEDATIVESMO=6;
ELSE IF S3BD1Q2E=5 THEN USFREQSEDATIVESMO=2.5;
ELSE IF S3BD1Q2E=6 THEN USFREQSEDATIVESMO=1;
ELSE IF S3BD1Q2E=7 THEN USFREQSEDATIVESMO=0.75;
ELSE IF S3BD1Q2E=8 THEN USFREQSEDATIVESMO=0.375;
ELSE IF S3BD1Q2E=9 THEN USFREQSEDATIVESMO=0.17;
ELSE IF S3BD1Q2E=10 THEN USFREQSEDATIVESMO=0.083;
/* usual frequency of seadtives assumption per month */
run;
ods graphics on;
/* ODS graphics turned on to manage the output and displays in HTML */
proc surveyselect data=new out=traintest seed = 123
samprate=0.7 method=srs outall;
run;
/* split data randomly into training data consisting of 70% of the total observations ()
test dataset consisting of the other 30% of the observations respectively)*/
/* data=new specifies the name of the managed input data set */
/* out equals the name of the randomly split output dataset, called traintest */
/* seed option to specify a random number seed to ensure that the data are randomly split the same way
if the code being run again */
/* samprate command split the input data set so that 70% of the observations
are being designated as training observations (the remaining 30% are being designated as test observations
respectively) */
/* method=srs specifies that the data are to be split using simple random sampling */
/* outall option includes, both the training and test observations in a single
output dataset which has a new variable called "selected", to indicate if an observation belongs
to the training set, or the test set */
proc glmselect data=traintest plots=all seed=123;
partition ROLE=selected(train='1' test='0');
/* glmselect procedure to test the lasso multiple regression w/ Least Angled Regression algorithm k=10 fold
validation
glmselect procedure standardize the predictor variables, so that they all have a mean equal to 0 and a
standard deviation equal to 1,
which places them all on the same scale
data=traintest to use the randomly split dataset
plots=all option to require that all plots associated w/ the lasso regression being printed
seed option to allow to specify a random number seed, being used in the cross-validation process
partition command to assign each observation a role, based on the variable called selected,
indicating if the observation is a training or test observation.
Observations with a value of 1 on the selected variable are assigned the role of training observation
(observations with a value of 0, are assigned the role of test observation respectively) */
model NUMALCMO_EST = MAJORDEPLIFE WhiteGroup BlackGroup
NamericaGroup AsianGroup HispanicGroup OPIOIDSRegularConsumer S3BD3Q2A
CANNABISRegularConsumer S3BD5Q2A SEDATIVESRegularConsumer S3BD1Q2A TRANQUILIZERSRegularConsumer
S3BD2Q2A COCAINERegularConsumer S3BD6Q2A TABLIFEDX LifetimeAlcAbuseDepy USFREQMO
NUMCIGMO_EST USFREQALCMO S3BD1Q2E
USFREQSEDATIVESMO/selection=lar(choose=cv stop=none) cvmethod=random(10);
/* model command to specify the regression model for which the response variable,
NUMALCMO_EST, is equal to the list of the 14 candidate predictor variables */
/* selection option to tell which method to use to compute the parameters for variable selection */
/*Least Angled Regression algorithm is being used */
/* choose=cv option to use cross validation for choosing the final statistical model */
/* stop=none to guarantee the model doesn't stop running until each of the candidate predictor
variables is being tested */
/* cvmethod=random(10) to specify a K-fold cross-validation method with ten randomly selected folds
is being used */
run;

0 notes
Text
LIBNAME mydata "/courses/d1406ae5ba27fe300 " access=readonly;
DATA new; set mydata.nesarc_pds;
IF S1Q6A eq 1 THEN incomegroup=.;
ELSE IF S1Q6A LE 3 THEN education=1;
ELSE IF S1Q6A LE 5 THEN education=2;
ELSE IF S1Q6A LE 7 THEN education=3;
ELSE IF S1Q6A GT 10 THEN education=3;
PROC SORT; by IDNUM;
PROC CORR; VAR S1Q10B S1Q11B;
RUN;
The CORR Procedure2 Variables:S1Q10B S1Q11BSimple StatisticsVariableNMeanStd DevSumMinimumMaximumS1Q10B430936.757504.40665291201017.00000S1Q11B430939.421414.843084059971.0000021.00000Pearson Correlation Coefficients, N = 43093
Prob > |r| under H0: Rho=0 S1Q10BS1Q11BS1Q10B
1.00000
0.67157
<.0001S1Q11B
0.67157
<.0001
1.00000
both education level is correlated to individual income and household income as shown by the data above. this is because the p value is less than .0001
0 notes
Text
Tarea semana 2
Mi primer programa Python
**** PROGRAMA ****
import pandas
import numpy
data = pandas.read_csv('nesarc_pds.csv', low_memory=False)
data['ETHRACE2A'].dtype
print(len(data))
print(len(data.columns))
#ajustar variables numéricas
# var1 IN WORST PERIOD, EVER FIND IT DIFFICULT TO STOP BEING TENSE/NERVOUS/WORRIED
#data['S9Q33']= pandas.to_numeric(data['S9Q33'])
# var2 IN WORST PERIOD, FELT UNCOMFORTABLE ABOUT FEELING NERVOUS/ANXIOUS OR BY ANY OF THOSE THINGS GOING ON AT SAME TIME
#data['S9Q51']= pandas.to_numeric(data['S9Q51'])
# varA FIND IT HARD TO START OR WORK ON TASKS WHEN THERE IS NO ONE TO HELP
#data['S10Q1A11']= pandas.to_numeric(data['S10Q1A11'])
# varB EVER TROUBLE YOU OR CAUSE PROBLEMS AT WORK/SCHOOL OR WITH FAMILY/OTHER PEOPLE
#data['S10Q1B11']= pandas.to_numeric(data['S10Q1B11'])
# varC WHEN AROUND PEOPLE, OFTEN FEEL THAT YOU ARE BEING WATCHED OR STARED AT
#data['S10Q1A35']= pandas.to_numeric(data['SS10Q1A35'])
print('S9Q33 - se dificulta dejar de estar ansioso o preocupado')
print('Conteo')
var1 = data['S9Q33'].value_counts(sort=False)
print(var1)
print('Porcentaje')
var1_pct = data['S9Q33'].value_counts(sort = False, normalize = True)
print(var1_pct )
print('S9Q51 - se siente incómodo por la ansiedad o preocupación')
print('Conteo')
var2 = data['S9Q51'].value_counts(sort=False)
print(var2)
print('Porcentaje')
var2_pct = data['S9Q51'].value_counts(sort = False, normalize = True)
print(var2_pct )
print('S10Q1A11 - encuentra dificultad para iniciar tareas')
print('Conteo')
varA = data['S10Q1A11'].value_counts(sort=False)
print(varA)
print('Porcentaje')
varA_pct = data['S10Q1A11'].value_counts(sort = False, normalize = True)
print(varA_pct )
print('S10Q1B11 - preocupa causar problemas')
print('Conteo')
varB = data['S10Q1B11'].value_counts(sort=False)
print(varB)
print('Porcentaje')
varB_pct = data['S10Q1B11'].value_counts(sort = False, normalize = True)
print(varB_pct )
print('S10Q1A35- se siente mal cuando hay gente alrededor')
print('Conteo')
varC = data['S10Q1A35'].value_counts(sort=False)
print(varC)
print('Porcentaje')
varC_pct = data['S10Q1A35'].value_counts(sort = False, normalize = True)
print(varC_pct )
conteo = data.groupby('S9Q33').size()
print(conteo)
conteo_pct = data.groupby('S9Q33').size()*100/len('data')
print(conteo_pct)
**** DATOS QUE ARROJA EL PROGRAMA ****
43093
3010
S9Q33 - se dificulta dejar de estar ansioso o preocupado
Conteo
S9Q33
39607
1 2715
2 742
9 29
Name: count, dtype: int64
Porcentaje
S9Q33
0.919105
1 0.063003
2 0.017219
9 0.000673
Name: proportion, dtype: float64
S9Q51 - se siente incómodo por la ansiedad o preocupación
Conteo
S9Q51
40189
1 1687
9 40
2 1177
Name: count, dtype: int64
Porcentaje
S9Q51
0.932611
1 0.039148
9 0.000928
2 0.027313
Name: proportion, dtype: float64
S10Q1A11 - encuentra dificultad para iniciar tareas
Conteo
S10Q1A11
2 40203
9 1441
1 1449
Name: count, dtype: int64
Porcentaje
S10Q1A11
2 0.932936
9 0.033439
1 0.033625
Name: proportion, dtype: float64
S10Q1B11 - preocupa causar problemas
S10Q1A35- se siente mal cuando hay gente alrededor
DESCRIPCION DE LA INFORMACIÓN
Se creo el programa depurando los errores que arrojaban y entendiendo lo que se está queriendo obtener.
Para las variable involucradas se sacó el conteo y porcentaje respectivamente, cada variable detalla el area de interés.
Las primeras dos son por malestar en la persona y las 3 ultimas indican una posible causa del malestar, solo se mestran 3 en las tablas de frecuencia.
En todas las variables 1 es YES, 2 NO, 3 UNKNOWN
#python #datamanagement
0 notes
Text
Making my first program assigment
This is my code in python:
#-- coding: utf-8 --
"""
Spyder Editor
This is a temporary script file.
"""
import pandas as pd
import numpy as np
data = pd.read_csv('nesarc_pds.csv', low_memory=False)
print(len(data))
print("counts for S2AQ1 usual quantity for drank at least 1 alcoholic drink in life by:"
"1=yes and 2=no")
c1=data['S2AQ1'].value_counts(sort=False, dropna=False)
print(c1)
print("percentages for S2AQ1 usual quantity for drank at least 1 alcoholic drink in life by:"
"1=yes and 2=no")
p1=data['S2AQ1'].value_counts(sort=False,dropna=False, normalize=True)
print(p1)
print("counts for S2AQ2 usual quantity for drank at least 12 alcoholic drink in last 12 months by:"
"1=yes, 2=no and 9=unknown")
c2=data['S2AQ2'].value_counts(sort=False, dropna=False)
print(c2)
print("percentage for S2AQ2 usual quantity for drank at least 12 alcoholic drink in last 12 months by:"
"1=yes, 2=no and 9=unknown")
p2=data['S2AQ2'].value_counts(sort=False, dropna=False, normalize=True)
print(p2)
print("counts for CONSUMER usual quatity the drinkins status categorizaded by:"
"1=Current drinker, 2=Ex-drinker and 3=Lifetime Abstainer")
c3=data['CONSUMER'].value_counts(sort=False, dropna=False)
print(c3)
print("percentages for CONSUMER usual quatity the drinkins status categorizaded by:"
"1=Current drinker, 2=Ex-drinker and 3=Lifetime Abstainer")
p3=data['CONSUMER'].value_counts(sort=False,normalize=True, dropna=False)
print(p3)
#As a subcollection I choose to work with people who are between 18 and 28 years old and they are currently drinkers
sub1=data[(data["AGE"]>=18) & (data["AGE"]<28) & (data["CONSUMER"]==1)]
sub2=sub1.copy()
#Now we can see if my sample its correctly
print("Counts for Age")
c4=sub2["AGE"].value_counts(sort=False)
print(c4)
print("Percentages for Age")
c5=sub2["AGE"].value_counts(sort=False, normalize=True)
print(c5)
print("Counts for CONSUMER")
c6=sub2["CONSUMER"].value_counts(sort=False)
print(c6)
print("Percentages for CONSUMER")
c7=sub2["CONSUMER"].value_counts(sort=False, normalize=True)
print("So we can see that just 5047 of the 43093 individuals are currently consumers and they are between 18 and 28 years old")
And his output is:
43093
counts for S2AQ1 usual quantity for drank at least 1 alcoholic drink in life by:1=yes and 2=no
S2AQ1
2 8266
1 34827
Name: count, dtype: int64
percentages for S2AQ1 usual quantity for drank at least 1 alcoholic drink in life by:1=yes and 2=no
S2AQ1
2 0.191818
1 0.808182
Name: proportion, dtype: float64
counts for S2AQ2 usual quantity for drank at least 12 alcoholic drink in last 12 months by:1=yes, 2=no and 9=unknown
S2AQ2
2 22225
1 20836
9 32
Name: count, dtype: int64
percentage for S2AQ2 usual quantity for drank at least 12 alcoholic drink in last 12 months by:1=yes, 2=no and 9=unknown
S2AQ2
2 0.515745
1 0.483512
9 0.000743
Name: proportion, dtype: float64
counts for CONSUMER usual quatity the drinkins status categorizaded by:1=Current drinker, 2=Ex-drinker and 3=Lifetime Abstainer
CONSUMER
3 8266
1 26946
2 7881
Name: count, dtype: int64
percentages for CONSUMER usual quatity the drinkins status categorizaded by:1=Current drinker, 2=Ex-drinker and 3=Lifetime Abstainer
CONSUMER
3 0.191818
1 0.625299
2 0.182884
Name: proportion, dtype: float64
Counts for Age
AGE
19 465
18 401
21 560
25 488
22 531
20 476
24 582
26 476
23 536
27 532
Name: count, dtype: int64
Percentages for Age
AGE
19 0.092134
18 0.079453
21 0.110957
25 0.096691
22 0.105211
20 0.094313
24 0.115316
26 0.094313
23 0.106202
27 0.105409
Name: proportion, dtype: float64
Counts for CONSUMER
CONSUMER
1 5047
Name: count, dtype: int64
Percentages for CONSUMER
So we can see that just 5047 of the 43093 individuals are currently consumers and they are between 18 and 28 years old
0 notes
Text
Alcohol intake and hypertension Data Visualisation
1. The program used for Visualisation

2. Data visualisation

Figure 1. Frequency of drinking status among participants (n=43093) in Nesarc study data

Figure 2. Frequency of drinking 12 alcoholic drinks in the last 12 months among participants (n=43093) in Nesarc study data. 1.0: Yes, 2.0: No, nan: Unknown.

Figure 3. Frequency of self-reported hypertension in the last 12 months among participants (n=43093) in Nesarc study data. 1.0: Yes, 2.0: No, nan: Unknown.

Figure 4: Frequency of alcoholic drink intake in the number of days in a month in the last 12 months among participants (n=43093) in Nesarc study data

Figure 5. Bar plot of drinking status and self-reported hypertension among participants in the last 12 months (n=43093) in Nesarc study data. 1.0: Yes, 2.0: No, nan: Unknown.
3. Results Interpretation
Figure 1 shows the frequency of drinking status among the 43,093 participants. The majority were current drinkers (62.5%), followed by lifetime abstainers (19.2%) and then ex-drinkers (18.3%).
Figure 2 displays the frequency of consuming alcoholic drinks in the past 12 months. Slightly less than half (48.3%) reported drinking alcohol in the past year, while 51.6% did not drink and 0.1% had unknown drinking status.
Figure 3 presents the frequency of self-reported hypertension diagnosis. The majority (76.2%) answered no, 21.2% answered yes, and 2.6% were unknown.
Figure 4 illustrates the frequency of days per month that alcohol was consumed over the past year among those who drank. The most common frequencies were 2.5 days/month (8.3%), 1 day/month (6.2%), and 0.38 days/month (7.4%).
Figure 5 compares the drinking status of alcohol between participants with and without self-reported hypertension. Current drinking was less frequent among those with hypertension (57.7%) versus those without (63.9%). Lifetime abstaining was more common for hypertension patients (22.5%) than non-hypertensive participants (18.5%).
In summary, the bar charts demonstrate that current drinking was most prevalent, with the majority of drinkers consuming alcohol about 1-3 days per month. Hypertension was self-reported in 21% of respondents and was less associated with current drinking than non-drinking status.
0 notes
Text
Running My First Program
I worked with a subset of the NESCAR dataset and analyzed the following three variables:
Buildingtype (appartment buildings only with an ordinal value (higher means more appartments))
Number of children ages 1-4 in household
Number of children ages under 18 in household
My program
import pandas
import numpy
data = pandas.read_csv('nesarc_pds.csv', low_memory=False)
print('-------------------------------------------------------------------------------------------------------------------')
print('Dataset used: nesarc - National Epidemiologic Survey of Drug Use and Health')
print('number of observations (rows)')
print (len(data)) # number of observations (rows)
print('number of variables (columns)')
print (len(data.columns)) # number of variables (columns)
print('')
print('Creating a subset with appartment building inhabitants only')
#subset data to appartments only
sub1=data[(data['BUILDTYP']>=4) & (data['BUILDTYP']<=9)]
#make a copy of my new subsetted data
sub_appartment = sub1.copy()
print('number of observations (rows)')
print (len(sub_appartment)) # number of observations (rows)
print('number of variables (columns)')
print (len(sub_appartment.columns)) # number of variables (columns)
print('')
print('-------------------------------------------------------------------------------------------------------------------')
counts and percentages (i.e. frequency distributions) for each variable
print('counts for BUILDTYP - TYPE OF BUILDING FOR HOUSEHOLD')
c1 = sub_appartment['BUILDTYP'].value_counts(sort=True, dropna=False)
print (c1)
print('percentages for BUILDTYP - TYPE OF BUILDING FOR HOUSEHOLD')
p1 = sub_appartment['BUILDTYP'].value_counts(sort=True, normalize=True)
print (p1)
print ('counts for CHLD1_4 - NUMBER OF CHILDREN AGES 1 THROUGH 4 IN HOUSEHOLD')
c2 = sub_appartment['CHLD1_4'].value_counts(sort=True, dropna=False)
print(c2)
print ('percentages for CHLD1_4 - NUMBER OF CHILDREN AGES 1 THROUGH 4 IN HOUSEHOLD')
p2 = sub_appartment['CHLD1_4'].value_counts(sort=True, normalize=True)
print (p2)
print ('counts for CHLD0_17 - NUMBER OF CHILDREN UNDER AGE 18 IN HOUSEHOLD')
c3 = sub_appartment['CHLD0_17'].value_counts(sort=True, dropna=False)
print(c3)
print ('percentages for CHLD0_17 - NUMBER OF CHILDREN UNDER AGE 18 IN HOUSEHOLD')
p3 = sub_appartment['CHLD0_17'].value_counts(sort=True, normalize=True)
print (p3)
Output
Dataset used: nesarc - National Epidemiologic Survey of Drug Use and Health
number of observations (rows)
43093
number of variables (columns)
3010
Creating a subset with appartment building inhabitants only
number of observations (rows)
11349
number of variables (columns)
3010
initial format of used variables
int64
int64
int64
counts for BUILDTYP - TYPE OF BUILDING FOR HOUSEHOLD
7 1775
9 1977
6 2133
8 1418
5 2131
4 1915
Name: BUILDTYP, dtype: int64
percentages for BUILDTYP - TYPE OF BUILDING FOR HOUSEHOLD
7 0.156401
9 0.174200
6 0.187946
8 0.124945
5 0.187770
4 0.168737
Name: BUILDTYP, dtype: float64
counts for CHLD1_4 - NUMBER OF CHILDREN AGES 1 THROUGH 4 IN HOUSEHOLD
0 9737
1 1252
2 321
3 37
4 2
Name: CHLD1_4, dtype: int64
percentages for CHLD1_4 - NUMBER OF CHILDREN AGES 1 THROUGH 4 IN HOUSEHOLD
0 0.857961
1 0.110318
2 0.028284
3 0.003260
4 0.000176
Name: CHLD1_4, dtype: float64
counts for CHLD0_17 - NUMBER OF CHILDREN UNDER AGE 18 IN HOUSEHOLD
0 7659
1 1728
3 514
2 1206
4 166
6 19
5 45
7 5
8 6
15 1
Name: CHLD0_17, dtype: int64
percentages for CHLD0_17 - NUMBER OF CHILDREN UNDER AGE 18 IN HOUSEHOLD
0 0.674861
1 0.152260
3 0.045290
2 0.106265
4 0.014627
6 0.001674
5 0.003965
7 0.000441
8 0.000529
15 0.000088
Name: CHLD0_17, dtype: float64
Summary
The Data shows that most households were in the buildingtype category 6 (buildings with 5 to 9 appartments) closely followed by category 5 (buildings with 3 to 4 appartments).
By far the most households (85.8%) had no child (1-4 years) and (67.5) no child below 18 years. In only one houshold there were 15 children.
There were no entries with missing data on children. For the buildingtype, the missing entries were filtered when the subset was created.
0 notes
Text
Assignment 2
The Code :
import pandas as pd
import numpy as np
#1 = yes , 2 = No , 9 = Unknown
data = pd.read_csv('~/Downloads/nesarc_pds.csv',low_memory = False)
#after drinking heavily than usual
c1 = data["S9Q10"].value_counts(sort = False, dropna=False)
p1 = data["S9Q10"].value_counts(sort = False, normalize = True, dropna=False)
#as after effect of drinking
c2 = data["S9Q11"].value_counts(sort = False, dropna=False)
p2 = data["S9Q11"].value_counts(sort = False, normalize = True, dropna=False)
#continued feeling tensed after stop drinking
c5 = data["S9Q14GR"].value_counts(sort = False, dropna=False)
p5 = data["S9Q14GR"].value_counts(sort = False, normalize = True, dropna=False)
#show in dataframes
S9Q10 = pd.concat([c1, p1], axis=1, keys=['Count', 'Percentage'])
S9Q10['Cumulative_Count'] = S9Q10['Count'].cumsum()
S9Q10['Cumulative_Percentage'] = S9Q10['Percentage'].cumsum()
S9Q11 = pd.concat([c2, p2], axis=1, keys=['Count', 'Percentage'])
S9Q11['Cumulative_Count'] = S9Q11['Count'].cumsum()
S9Q11['Cumulative_Percentage'] = S9Q11['Percentage'].cumsum()
S9Q11
S9Q14GR = pd.concat([c5, p5], axis=1, keys=['Count', 'Percentage'])
S9Q14GR['Cumulative_Count'] = S9Q14GR['Count'].cumsum()
S9Q14GR['Cumulative_Percentage'] = S9Q14GR['Percentage'].cumsum()
S9Q14GR
Result :



Explanation :
I did not filter the data or add in a sub question as I want to understand how drinking affects generalized anxiety to everyone. From the data I got from python, I see that not many people get generalized anxiety because of drinking. I have used the dropna = False code to handle the missing data so that it does not remove missing data. Hence the blank index in the result shows the missing data.
Blank index : The one who did not meet symptom criteria
1: Yes
2:No
9:Unknown
The first question (S9Q10) : Episode began after drinking heavily than usual
Out of 43093 people who have answered, about 2740 have answered No and 100 has said yes.
The 2nd Question (S9Q10) : Episode began after experiencing bad after effects of drinking
Out of 43093 people who have answered, about 2768 have answered No and 71 has said yes.
The 3rd Question (S9Q14GR) : Episode continued after stop drinking
Out of 43093 people who have answered, about 1393 people have answered No and 23 has answered yes.
0 notes
Text
second project assignment
here is my submission for the project
# Step 1: Import libraries
import pandas
import numpy#Step 2: Import/Load/Read data file
data = pandas.read_csv('nesarc_pds.csv', low_memory=False)#Convert datatypes to numeric
data['S1Q10B'] = pandas.to_numeric(data['S1Q10B'],errors='coerce') #1
data['S12Q1'] = pandas.to_numeric(data['S12Q1'],errors='coerce') #2
data['S12Q2B10'] = pandas.to_numeric(data['S12Q2B10'],errors='coerce') #3
data['S12Q2B12'] = pandas.to_numeric(data['S12Q2B12'],errors='coerce') #4#Print Header and Research Question
print("""
WEEK 2 ASSIGNMENT: WRITING YOUR FIRST PROGRAM
BY: ASHLEY R. Intended Research Question: In the last 12 months, is a persons income level associated with destructive gambling habits or patterns? """)#Show number of rows and columns.
print("Total Number of Observations (Rows) in NESARC file")
print (len(data))
print("Total Number of Variables (Columns) in NESARC file")
print(len(data.columns)) #Counts and Percentages of all related variables#identify code meanings (create legend)
print ("""
for all variable codes, excluding categories:
1 = yes;
2 = no;
9 = unknown;
BL/NaN = NA, never or unknown if ever gambled 5+ times in one year
==============================================================""")#1 - personal income
print("""
S1Q10B - total personal income in the last 12 months, category""")
print ("""
Code Counts""")
c1 = data["S1Q10B"].value_counts(sort=False, dropna=False).sort_index(ascending= True)
print (c1)print("""
Code Percentages""")
p1 = data["S1Q10B"].value_counts(sort=False, normalize=True, dropna=False).sort_index(ascending= True)
print (p1)#2 - Ever gambled regualarly
print("""
S12Q1 - Ever gambled 5+ times in any one year""")
print ("""
Code Counts""")
c2 = data["S12Q1"].value_counts(sort=False, dropna=False).sort_index(ascending= True)
print (c2)print("""
Code Percentages""")
p2 = data["S12Q1"].value_counts(sort=False, normalize=True, dropna=False).sort_index(ascending= True)
print (p2)#3 - Financial Trouble - living expenses - last 12 months
print("""
S12Q2B10 - Ever have such financial trouble that you had to get help with living expenses from family, friends, or welfare in the last 12 months""")
print ("""
Code Counts""")
c3 = data["S12Q2B10"].value_counts(sort=False, dropna=False).sort_index(ascending= True)
print (c3)print("""
Code Percentages""")
p3 = data["S12Q2B10"].value_counts(sort=False, normalize=True, dropna=False).sort_index(ascending= True)
print (p3)#4 - Stealing and bad checks - last 12 months
print("""
S12Q2B12 - Ever raise gambling money by writing a bad check, signing someone else's name to a check, stealing, cashing someone else's check or in some other illegal way in the last 12 months""")
print ("""
Code Counts""")
c4 = data["S12Q2B12"].value_counts(sort=False, dropna=False).sort_index(ascending= True)
print (c4)print("""
Code Percentages""")
p4 = data["S12Q2B12"].value_counts(sort=False, normalize=True, dropna=False).sort_index(ascending= True)
print (p4)Output:
WEEK 2 ASSIGNMENT: WRITING YOUR FIRST PROGRAM
BY: ASHLEY R. Intended Research Question: In the last 12 months, is a persons income level associated with destructive gambling habits or patterns?Total Number of Observations (Rows) in NESARC file
43093
Total Number of Variables (Columns) in NESARC file
3008for all variable codes, excluding categories:
1 = yes;
2 = no;
9 = unknown;
BL/NaN = NA, never or unknown if ever gambled 5+ times in one year
============================================================== S1Q10B - total personal income in the last 12 months, categoryCode Counts
0 2462
1 3571
2 3823
3 2002
4 3669
5 1634
6 3940
7 3887
8 3085
9 3003
10 2351
11 3291
12 2059
13 1328
14 857
15 521
16 290
17 1320
Name: S1Q10B, dtype: int64Code Percentages
0 0.057132
1 0.082867
2 0.088715
3 0.046458
4 0.085141
5 0.037918
6 0.091430
7 0.090200
8 0.071589
9 0.069686
10 0.054556
11 0.076370
12 0.047780
13 0.030817
14 0.019887
15 0.012090
16 0.006730
17 0.030631
Name: S1Q10B, dtype: float64 S12Q1 - Ever gambled 5+ times in any one yearCode Counts
1 11153
2 30885
9 1055
Name: S12Q1, dtype: int64Code Percentages
1 0.258812
2 0.716706
9 0.024482
Name: S12Q1, dtype: float64 S12Q2B10 - Ever have such financial trouble that you had to get help with living expenses from family, friends, or welfare in the last 12 monthsCode Counts
1.0 49
2.0 11085
9.0 19
NaN 31940
Name: S12Q2B10, dtype: int64Code Percentages
1.0 0.001137
2.0 0.257234
9.0 0.000441
NaN 0.741188
Name: S12Q2B10, dtype: float64 S12Q2B12 - Ever raise gambling money by writing a bad check, signing someone else's name to a check, stealing, cashing someone else's check or in some other illegal way in the last 12 monthsCode Counts
1.0 11
2.0 11122
9.0 20
NaN 31940
Name: S12Q2B12, dtype: int64Code Percentages
1.0 0.000255
2.0 0.258093
9.0 0.000464
NaN 0.741188
Name: S12Q2B12, dtype: float64
0 notes
Text
Running Your First Program
I wrote my first program, to get to the bottom of my research question:
#-- coding: utf-8 --
"""
Created on Wed Jun 14 11:18:20 2023
@author:
"""
import pandas
import numpy
print("start import")
data = pandas.read_csv('nesarc_pds.csv', low_memory=False)
print("import done")
#upper-case all Dataframe column names --> unification
data.colums = map(str.upper, data.columns)
#bug fix for display formats to avoid run time errors
pandas.set_option('display.float_format', lambda x:'%f'%x)
print (len(data)) #number of observations (rows)
print (len(data.columns)) # number of variables (columns)
#checking the format of your variables
#setting variables you will be working with to numeric
data['S2DQ1'] = pandas.to_numeric(data['S2DQ1']) #Blood/Natural Father
data['S2DQ2'] = pandas.to_numeric(data['S2DQ2']) #Blood/Natural Mother
data['S2BQ3A'] = pandas.to_numeric(data['S2BQ3A'], errors='coerce') #Age at
data['S3CQ14A3'] = pandas.to_numeric(data['S3CQ14A3'], errors='coerce')
#Blood/Natural Father was alcoholic
print("number blood/natural father was alcoholic")
c1 = data['S2DQ1'].value_counts(sort=False).sort_index()
print (c1)
print("percentage blood/natural father was alcoholic")
p1 = data['S2DQ1'].value_counts(sort=False, normalize=True).sort_index()
print (p1)
#Blood/Natural Mother was alcoholic
print("number blood/natural mother was alcoholic")
c2 = data['S2DQ2'].value_counts(sort=False).sort_index()
print(c2)
print("percentage blood/natural mother was alcoholic")
p2 = data['S2DQ2'].value_counts(sort=False, normalize=True).sort_index()
print (p2)
#Age at onset of alcohol abuse --> Age is irrelevant; number of people with or without alcohol abuse is calculated
#1 = alcohol abuse; 2 = no alcohol abuse; 9 = unknown
print("number alcohol abuse")
#replace age with 1, 2 or 9
data['S2BQ3A'] = data['S2BQ3A'].replace({5: 1, 6: 1, 7: 1, 8: 1, 9: 1, 10: 1, 11: 1, 12: 1, 13: 1, 14: 1, 15: 1, 16: 1, 17: 1, 18: 1, 19: 1, 20: 1, 21: 1, 22: 1, 23: 1, 24: 1, 25: 1, 26: 1, 27: 1, 28: 1, 29: 1, 30: 1, 31: 1, 32: 1, 33: 1, 34: 1, 35: 1, 36: 1, 37: 1, 38: 1, 39: 1, 40: 1, 41: 1, 42: 1, 43: 1, 44: 1, 45: 1, 46: 1, 47: 1, 48: 1, 49: 1, 50: 1, 51: 1, 52: 1, 53: 1, 54: 1, 55: 1, 56: 1, 57: 1, 58: 1, 59: 1, 60: 1, 61: 1, 62: 1, 63: 1, 64: 1, 65: 1, 66: 1, 67: 1, 68: 1, 69: 1, 70: 1, 71: 1, 72: 1, 73: 1, 74: 1, 75: 1, 76: 1, 77: 1, 78: 1, 79: 1, 80: 1, 81: 1, 82: 1, 83: 1, 84: 1, 85: 1, 86: 1, 87: 1, 99: 9})
data['S2BQ3A'].fillna(2, inplace=True)
c3 = data['S2BQ3A'].value_counts().sort_index()
print(c3)
print("percentage alcohol abuse")
p3 = data['S2BQ3A'].value_counts(sort=False, normalize=True).sort_index()
print (p3)
#ever used Drugs/medicine in higher amount or longer period
print("number drugs/medicine abuse")
#blanks are filled with 9 (unknown)
data['S3CQ14A3'].fillna(9, inplace=True)
c4 = data['S3CQ14A3'].value_counts(sort=False).sort_index()
print(c4)
print("percentage drugs/medicine abuse")
p4 = data['S3CQ14A3'].value_counts(sort=False, normalize=True).sort_index()
print (p4)
the output of my program is the following:
start import
import done
43093
3010
number blood/natural father was alcoholic
1 8124
2 32445
9 2524
Name: S2DQ1, dtype: int64
percentage blood/natural father was alcoholic
1 0.188522
2 0.752907
9 0.058571
Name: S2DQ1, dtype: float64
number blood/natural mother was alcoholic
1 2311
2 39553
9 1229
Name: S2DQ2, dtype: int64
percentage blood/natural motherwas alcoholic
1 0.053628
2 0.917852
9 0.028520
Name: S2DQ2, dtype: float64
number alcohol abuse
1.000000 10293
2.000000 31923
9.000000 877
Name: S2BQ3A, dtype: int64
percentage alcohol abuse
1.000000 0.238855
2.000000 0.740793
9.000000 0.020351
Name: S2BQ3A, dtype: float64
number drugs/medicine abuse
1.000000 1244
2.000000 7832
9.000000 34017
Name: S3CQ14A3, dtype: int64
percentage drugs/medicine abuse
1.000000 0.028868
2.000000 0.181746
9.000000 0.789386
Name: S3CQ14A3, dtype: float64
the output of the program shows, that the percentage of people with alcoholic fathers compared to mothers is rather high (19% fathers and 5% mothers). even though the percentage of "unknown" is higher with the fathers.
one can also clearly see that the percentage of alcohol abuse (24%) is near to the percentage of fathers being alcoholic (19%)
for medicine and drug abuse the number of "unknown" is significantly higher than for the alcohol questions. The percentage of known medicine or drug abuse is rather low (3%)
0 notes
Text
Data Management and Visualization assignment week 1


Developing a Research Question and creating a personal Codebook
Introduction
After reviewing the codebook of the U.S. National Epidemiological Survey on Alcohol and Related Conditions (NESARC), following a survey of 43093 US citizens above 18 years, this survey was designed to explain the magnitude of alcohol use and psychiatric disorders, I found myself particularly interested in the crack/cocaine use and the disorder of it. And I also intend to examine the means of association between Crack/cocaine use and depression, as well as general anxiety disorders diagnosed in the last 12 months of this survey. Among people aged 12 or older in 2021, 1.7% (or about 4.8 million people) reported using cocaine in the past 12 months [1] and only in 2021, approximately 24,486 people died from an overdose involving cocaine [2] so it's interesting to see the correlation therein. It is common knowledge that there is a big relationship between the abuse of alcohol and mental disorder, therefore it is only necessary to see whether the frequent use of cocaine co-occurs with depression and general anxiety
RESEARCH Question.
Is the use of crack/cocaine associated with major depression and general anxiety disorder diagnoses in the last 12 months?
Hypothesis
Though a lot of research has gone into comparing the use of Crack/cocaine with general anxiety and depression, According to a report by the National Institute for Drug Abuse states that users of Large amounts of cocaine may intensify their high which can also lead to bizarre, erratic, and violent behavior. Some cocaine users report feelings of restlessness, irritability, anxiety, panic, paranoia, etc,[3] these findings make it easy to deduce the correlation between the use of cocaine and depression. My personal belief is that cocaine use increases the likelihood of depression symptoms and anxiety disorders, but not as significantly as cannabis abuse/dependence, which could cause mental disorders like the ones mentioned above.
NESARC Codebook Sections and Variables
After going through the NERASC code, I First, had a look at the unique numbers (IDNUM) to understand the number of samples that were working with and compared it to a variable of a smaller number (AGE) to have a better view to work with from background information in section 1 to make my findings a lot more reliable. Next the first question topic I chose from the drug use section (SECTION 3B) to include information like the percentage of people who ever used Cocaine (S3BQ1A6), The age they first used cocaine (S3BD6Q2A) as well as the period of last use of cocaine in the last 12 months/ prior to last 12 months and both periods (S3BD6Q2B), I also checked the frequency of using cocaine in the past 12 months (S3BD6Q2C), compared it with the number of days since their most recent cocaine use (S3BD6Q2DR) and checked it against the age when they began to use cocaine(S3BD6Q2F). As far as the second question is concerned, I selected the variable of non-hierarchical major depression diagnosis, in the last 12 months (MAJORDEP12) and the variable of non-hierarchical generalized anxiety diagnosis, in the last 12 months (GENAXDX12), which are included in the diagnoses section (SECTION 14).
Literature review
Taking into account the literature review I performed, using Google Scholar, I found several academic studies and research based on the relationship between cocaine use, depression, and anxiety there was a moderate association between involvement with its use in the past 12 months and the prevalence of affective and anxiety disorders.
In a sample of 298 cocaine abusers seeking inpatient treatment, rates of psychiatric disorders were determined by means of the Schedule for Affective Disorders and Research Diagnostic Criteria. Overall, 55.7% met current and 73.5% met lifetime criteria for a psychiatric disorder other than a substance use disorder. In common with previous reports from clinical samples of cocaine abusers, these overall rates were largely accounted for by major depression, minor bipolar conditions (eg, hypomania, cyclothymic personality), anxiety disorders, antisocial personality, and a history of childhood attention deficit disorder. Affective disorders and alcoholism usually followed the onset of drug abuse, while anxiety disorders, antisocial personality, and attention deficit disorder typically preceded drug abuse. [4]. These associations did not remain significant after including demographics, neuroticism, and other drug use in multiple regressions. Cocaine use did not appear to be directly related to depression or anxiety when the account was taken of other drug use. However, the association between heavier involvement with the use and affective and anxiety disorders has implications for the treatment of persons with problematic use.
https://www.samhsa.gov/data/report/2021-nsduh-annual-national-report
https://nida.nih.gov/research-topics/trends-statistics/overdose-death-rates
https://nida.nih.gov/publications/research-reports/cocaine/references
https://jamanetwork.com/journals/jamapsychiatry/article-abstract/495193
0 notes
Text
Is environment associated with fear in people's heart?
After reading the code book for the NESARC study, I am curious about people's fear in their heart. There are some variables regarding to our living environment, including census, family members, etc.
Through these information, I wish I can find the relevance between surrounding and our heart. I'm going to analyze the data into three parts: family and friends, past experience, and substantial things.
The following are variables I will probably use.
The information about fear or any feeling that is uncomfortable
Tape Location 2908-2949
The information about environment where people live
42-42 CENDIV CENSUS DIVISION
2018 1. New England
6191 2. Middle Atlantic
6430 3. East North Central
2561 4. West North Central
8665 5. South Atlantic
2658 6. East South Central
4832 7. West South Central
3046 8. Mountain
6692 9. Pacific
46-47 BUILDTYP TYPE OF BUILDING FOR HOUSEHOLD
2615 1. Mobile home
25506 2. Detached one-family house
2484 3. Attached one-family house
1915 4. Building with 2 apartments
2131 5. Building with 3 to 4 apartments
2133 6. Building with 5 to 9 apartments
1775 7. Building with 10 to 19 apartments
1418 8. Building with 20 to 49 apartments
1977 9. Building with 50+ apartments
15 10. "Other (boat, RV, etc.)"
6 11. One-family house (unspecified 2 or 3)
153 12. Apartment (unspecified 4-9)
965 99. Unknown building type
48-49 NUMPERS NUMBER OF PERSONS IN HOUSEHOLD
43093 1-17. Persons
50-51 NUMPER18 NUMBER OF PERSONS 18 YEARS AND OLDER IN HOUSEHOLD
43093 1-10. Persons
52-53 NUMREL NUMBER OF RELATED PERSONS IN HOUSEHOLD, INCLUDING SAMPLE PERSON
43093 1-17. Persons
54-54 NUMREL18 NUMBER OF RELATED PERSONS 18 YEARS AND OLDER IN HOUSEHOLD,
INCLUDING SAMPLE PERSON
43093 1-9. Persons
55-55 CHLD0 NUMBER OF CHILDREN UNDER AGE 1 IN HOUSEHOLD
42380 0. None
713 1-6. Number of children
56-56 CHLD1_4 NUMBER OF CHILDREN AGES 1 THROUGH 4 IN HOUSEHOLD
37127 0. None
5966 1-5. Number of children
57-57 CHLD5_12 NUMBER OF CHILDREN AGES 5 THROUGH 12 IN HOUSEHOLD
33424 0. None
9669 1-8. Number of children
58-58 CHLD13_15 NUMBER OF CHILDREN AGES 13 THROUGH 15 IN HOUSEHOLD
38535 0. None
4558 1-4. Number of children
59-59 CHLD16_17 NUMBER OF CHILDREN AGES 16 OR 17 IN HOUSEHOLD
39949 0. None
3144 1-3. Number of children
60-61 CHLD0_17 NUMBER OF CHILDREN UNDER AGE 18 IN HOUSEHOLD
26940 0. None
16153 1-15. Number of children
2908-2949
1 note
·
View note
Text
Running program assignment -week 2
importpandasimportnumpy
In [4]: print ("this my first experiment") this my first experiment
In [16]:data = pandas.read_csv("_c10361280c0613304594ab464c014f47_nesarc_pds.csv" , low_memory=False)
In [20]:print(data) print(len(data.columns)) Unnamed: 0 ETHRACE2A ETOTLCA2 IDNUM PSU STRATUM WEIGHT \ 0 0 5 1 4007 403 3928.613505 1 1 5 0.0014 2 6045 604 3638.691845 2 2 5 3 12042 1218 5779.032025 3 3 5 4 17099 1704 1071.754303 4 4 2 5 17099 1704 4986.952377 ... ... ... ... ... ... ... ... 43088 43088 1 43089 12010 1208 10477.240840 43089 43089 1 0.2237 43090 17099 1704 9014.746280 43090 43090 1 0.3785 43091 18094 1802 8079.917091 43091 43091 1 14.0831 43092 31035 3104 10367.259020 43092 43092 1 43093 17099 1704 9014.746280 CDAY CMON CYEAR ... SOLP12ABDEP HAL12ABDEP HALP12ABDEP \ 0 14 8 2001 ... 0 0 0 1 12 1 2002 ... 0 0 0 2 23 11 2001 ... 0 0 0 3 9 9 2001 ... 0 0 0 4 18 10 2001 ... 0 0 0 ... ... ... ... ... ... ... ... 43088 27 11 2001 ... 0 0 0 43089 30 10 2001 ... 0 0 0 43090 16 10 2001 ... 0 0 0 43091 26 9 2001 ... 3 0 3 43092 1 11 2001 ... 0 0 0 MAR12ABDEP MARP12ABDEP HER12ABDEP HERP12ABDEP OTHB12ABDEP \ 0 0 0 0 0 0 1 0 0 0 0 0 2 0 0 0 0 0 3 0 0 0 0 0 4 0 0 0 0 0 ... ... ... ... ... ... 43088 0 0 0 0 0 43089 0 0 0 0 0 43090 0 0 0 0 0 43091 0 3 0 0 0 43092 1 1 0 0 0 OTHBP12ABDEP NDSymptoms 0 0 NaN 1 0 NaN 2 0 NaN 3 0 NaN 4 0 NaN ... ... ... 43088 0 NaN 43089 0 NaN 43090 0 NaN 43091 0 NaN 43092 0 NaN [43093 rows x 3010 columns] 3010
In [24]:print(len(data.columns)) 3010
In [31]:data['S3AQ2A2'].dtype
Out[31]:dtype('O')
In [32]:data['S3AQ51'].dtype
Out[32]:dtype('O')
In [34]:print(data) Unnamed: 0 ETHRACE2A ETOTLCA2 IDNUM PSU STRATUM WEIGHT \ 0 0 5 1 4007 403 3928.613505 1 1 5 0.0014 2 6045 604 3638.691845 2 2 5 3 12042 1218 5779.032025 3 3 5 4 17099 1704 1071.754303 4 4 2 5 17099 1704 4986.952377 ... ... ... ... ... ... ... ... 43088 43088 1 43089 12010 1208 10477.240840 43089 43089 1 0.2237 43090 17099 1704 9014.746280 43090 43090 1 0.3785 43091 18094 1802 8079.917091 43091 43091 1 14.0831 43092 31035 3104 10367.259020 43092 43092 1 43093 17099 1704 9014.746280 CDAY CMON CYEAR ... SOLP12ABDEP HAL12ABDEP HALP12ABDEP \ 0 14 8 2001 ... 0 0 0 1 12 1 2002 ... 0 0 0 2 23 11 2001 ... 0 0 0 3 9 9 2001 ... 0 0 0 4 18 10 2001 ... 0 0 0 ... ... ... ... ... ... ... ... 43088 27 11 2001 ... 0 0 0 43089 30 10 2001 ... 0 0 0 43090 16 10 2001 ... 0 0 0 43091 26 9 2001 ... 3 0 3 43092 1 11 2001 ... 0 0 0 MAR12ABDEP MARP12ABDEP HER12ABDEP HERP12ABDEP OTHB12ABDEP \ 0 0 0 0 0 0 1 0 0 0 0 0 2 0 0 0 0 0 3 0 0 0 0 0 4 0 0 0 0 0 ... ... ... ... ... ... 43088 0 0 0 0 0 43089 0 0 0 0 0 43090 0 0 0 0 0 43091 0 3 0 0 0 43092 1 1 0 0 0 OTHBP12ABDEP NDSymptoms 0 0 NaN 1 0 NaN 2 0 NaN 3 0 NaN 4 0 NaN ... ... ... 43088 0 NaN 43089 0 NaN 43090 0 NaN 43091 0 NaN 43092 0 NaN [43093 rows x 3010 columns]
In [54]:data['TAB12MDX'] = pandas.to_numeric(data['TAB12MDX']) data['CHECK321'] = pandas.to_numeric(data['CHECK321']) data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1']) data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1']) data['AGE'] = pandas.to_numeric(data['AGE'])
In [54]:data['TAB12MDX'] = pandas.to_numeric(data['TAB12MDX']) data['CHECK321'] = pandas.to_numeric(data['CHECK321']) data['S3AQ3B1'] = pandas.to_numeric(data['S3AQ3B1']) data['S3AQ3C1'] = pandas.to_numeric(data['S3AQ3C1']) data['AGE'] = pandas.to_numeric(data['AGE'])
In [55]:data ['TAB12MDX'].dtype
Out[55]:dtype('int64')
In [56]:data ['CHECK321'].dtype
Out[56]:dtype('float64')
In [57]:data ['S3AQ3B1'].dtype
Out[57]:dtype('float64')
In [61]:q1=data.groupby('TAB12MDX').size()
In [62]:print(q1) TAB12MDX 0 38131 1 4962 dtype: int64
In [63]:sub1=data[(data['AGE']>=18) & (data['AGE']<=25) & (data['CHECK321']==1)]
In [65]:print (sub1) Unnamed: 0 ETHRACE2A ETOTLCA2 IDNUM PSU STRATUM WEIGHT \ 20 20 2 0.0099 21 36094 3616 1528.354757 76 76 5 0.2643 77 36094 3616 6172.249980 102 102 1 0.985 103 41097 4107 5515.974591 121 121 1 0.8888 122 31098 3109 4152.434010 135 135 1 0.017 136 12042 1218 8657.814391 ... ... ... ... ... ... ... ... 42940 42940 2 42941 42099 4205 1564.139226 42989 42989 5 0.0999 42990 6046 605 3486.254607 42997 42997 1 0.6602 42998 53099 5304 12918.162000 43087 43087 1 0.2641 43088 42037 4209 9663.995112 43090 43090 1 0.3785 43091 18094 1802 8079.917091 CDAY CMON CYEAR ... SOLP12ABDEP HAL12ABDEP HALP12ABDEP \ 20 2 11 2001 ... 0 0 0 76 13 3 2002 ... 0 0 0 102 27 10 2001 ... 0 0 1 121 23 9 2001 ... 0 0 0 135 11 12 2001 ... 0 0 0 ... ... ... ... ... ... ... ... 42940 7 2 2002 ... 0 0 0 42989 9 2 2002 ... 0 0 0 42997 14 2 2002 ... 0 0 0 43087 23 1 2002 ... 0 0 0 43090 16 10 2001 ... 0 0 0 MAR12ABDEP MARP12ABDEP HER12ABDEP HERP12ABDEP OTHB12ABDEP \ 20 0 0 0 0 0 76 0 0 0 0 0 102 0 1 0 0 0 121 0 0 0 0 0 135 0 1 0 0 0 ... ... ... ... ... ... 42940 0 0 0 0 0 42989 0 0 0 0 0 42997 0 1 0 0 0 43087 0 0 0 0 0 43090 0 0 0 0 0 OTHBP12ABDEP NDSymptoms 20 0 2.0 76 0 NaN 102 0 3.0 121 0 3.0 135 0 0.0 ... ... ... 42940 0 4.0 42989 0 NaN 42997 0 6.0 43087 0 2.0 43090 0 NaN [1706 rows x 3010 columns]
In [67]:q2 = data.groupby('TAB12MDX').size() * 100 / len(data)
In [68]:print (q2) TAB12MDX 0 88.485369 1 11.514631 dtype: float64
In [75]:c5 = sub2['AGE'].value_counts(sort=False)
In [76]:print (c5) 18 161 19 200 20 221 21 239 22 228 23 231 24 241 25 185 Name: AGE, dtype: int64
In [77]:p5 = sub2['AGE'].value_counts(sort=False, normalize=True)
In [78]:print (p5) 18 0.094373 19 0.117233 20 0.129543 21 0.140094 22 0.133646 23 0.135404 24 0.141266 25 0.108441 Name: AGE, dtype: float64
In [79]:c6 = sub2['S2AQ8A'].value_counts(sort=False)
In [80]:print (c6) 5 216 99 8 10 118 6 248 180 2 84 3 194 9 134 7 134 1 76 4 229 8 85 Name: S2AQ8A, dtype: int64
In [81]:pandas.set_option('display.float_format', lambda x:'%f'%x)
In [82]:print ('display.float_format') display.float_format
0 notes
Text
Developing a Research Question and Creating a Personal Code Book
Introduction:
I became particularly interested in cannabis use problems after reading the NESARC codebook, a study of approximately 43093 American adults (over the age of 18) intended to gauge the prevalence of alcohol use and mental illnesses. More specifically, I want to look at the patterns of correlation between cannabis usage and recent diagnoses of major depressive disorder and generalised anxiety disorder. Cannabis is currently the most extensively used narcotic in many nations, but it also has medical use. According to estimates, 10% of cannabis users develop a dependency on the drug [4]. It is interesting to learn whether often using cannabis is associated with depression and generalized anxiety because it is well known that mental problems and alcohol misuse are strongly correlated.
Research Question:
Has anyone ever been diagnosed with a serious depressive illness or a generalized anxiety condition after using cannabis?
Hypothesis:
Although various research have looked at concerns related to cannabis usage disorders, it is challenging to determine whether cannabis use causes mental diseases. At this point, it's important to distinguish cannabis usage from a more serious drug use (dependence/abuse). My own opinion is that cannabis usage raises the risk of anxiety and depressive symptoms, but not as much as cannabis misuse or dependency, which can result in mental illnesses like the ones listed above.
NESARC Codebook Sections and Variables:
After looking through NESARC codebook, firstly i decided to take into consideration the unique identification number ( IDNUM ) and the variable (AGE) from background information ( SECTION 1 ) of the sample, in order to make my findings more reliable. Furthermore for my first question topic I chose, from the drug / medicine use section ( SECTION 3B ), to include information like the percentage of people who ever used cannabis ( S3BQ1A5 ), as well as the period of this use -last 12 months / prior to last 12 months / both periods- ( S3BD5Q2B ) and the frequency of it when using the most ( S3BD5Q2E ). As far as the second topic is concerned, I selected the variable of non-hierarchical major depression diagnoses, in last 12 months ( MAJORDEP12 ) and the variable of non-hierarchical generalized anxiety diagnoses, in last 12 months ( GENAXDX12 ), which are included in the diagnoses section ( SECTION 14 ).
SECTION 1 variables : IDNUM , AGE
SECTION 3B variables : S3BQ1A5 , S3BD5Q2B , S3BD5Q2E
SECTION 14 variables : MAJORDEP12 , GENAXDX12







Literature review:
Taking into account the literature review I performed, using Google Scholar, I found several academic studies and researches based on the relationship between cannabis use, depression and anxiety there was a moderate association between involvement with cannabis use in the past 12 months and the prevalence of affective and anxiety disorders. Among those with DSM-IV cannabis dependence, 14 % had affective disorder symptoms, compared to 6 % of non-users; while 17 % met criteria for an anxiety disorder, compared to 5 % of non-users [4]. These associations did not remain significant after including demographics, neuroticism and other drug use in multiple regressions. Cannabis use did not appear to be directly related to depression or anxiety when account was taken of other drug use. However, the association between heavier involvement with cannabis use and affective and anxiety disorders has implications for the treatment of persons with problematic cannabis use [4].
References:
Degenhardt, L., Hall, W., & Lynskey, M. (2003). Exploring the association between cannabis use and depression. Addiction, 98(11), 1493-1504.
Hayatbakhsh, M. R., Najman, J. M., Jamrozik, K., Mamun, A. A., Alati, R., & Bor, W. (2007). Cannabis and anxiety and depression in young adults: a large prospective study. Journal of the American Academy of Child & Adolescent Psychiatry, 46(3), 408-417.
Degenhardt, L., Hall, W., & Lynskey, M. (2001). Alcohol, cannabis and tobacco use among Australians: a comparison of their associations with other drug use and use disorders, affective and anxiety disorders, and psychosis. Addiction, 96(11), 1603-1614.
Degenhardt, L., Hall, W., & Lynskey, M. (2001). The relationship between cannabis use, depression and anxiety among Australian adults: findings from the National Survey of Mental Health and Well-Being. Social psychiatry and psychiatric epidemiology, 36(5), 219-227.
1 note
·
View note
Text
Creating graphs for your data
print("ahmed")
ahmed
ahmed = [1,2,3,4,5,6,7,8,9]
print (ahmed[4])
5
ahmed hindi
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
import seaborn
read pickled data
data = pd.read_pickle('cleaned_data2.pickle')
data.shape
(43093, 12)
data.dtypes
marital object
age_1st_mar object
age int64
hispanich int64
indian int64
asian int64
black int64
HAWAIIAN int64
WHITE int64
how_mar_ended object
edu object
ETHNICITY object
dtype: object
data.head()
marita
l
age_1st_
mar
ag
e
hispani
ch
indi
an
asia
n
bla
ck
HAWAII
AN
WHI
TE
how_mar_e
nded edu ETHNIC
ITY
0
Never
Marrie
d
23 1 2 2 2 2 1
Comple
ted high
school
hispani
ch
1 Marrie
d 23 28 1 2 2 2 2 1
Comple
ted high
school
hispani
ch
2 Widow
ed 35 81 1 2 2 2 2 1 2 8 hispani
ch
3
Never
Marrie
d
18 1 2 2 2 2 1
Comple
ted high
school
hispani
ch
4 Marrie
d 22 36 2 2 2 1 2 2 bachelo
r's black
%matplotlib inline
barplot (count plot) for the marital status
# univariate bar graph for categorical variables
# First hange format from numeric to categorical
plt.figure(figsize=(15,5))
data["marital"] = data["marital"].astype('category')
seaborn.countplot(x="marital", data=data)
plt.xlabel('marital ')
barplot (count plot) for the education level
plt.figure(figsize=(18,8))
data["edu"] = data["edu"].astype('category')
seaborn.countplot(x="edu", data=data)
plt.xlabel('education ')
barplot (count plot) for the ETHNICITY .
plt.figure(figsize=(10,5))
data["ETHNICITY"] = data["ETHNICITY"].astype('category')
seaborn.countplot(x="ETHNICITY", data=data)
plt.xlabel('ETHNICITY ')
the distribution od the ages in the sample
plt.figure(figsize=(18,8))
seaborn.distplot(data["age"].dropna(), kde=False);
plt.xlabel('Age')
# plt.figure(figsize=(18,8))
# seaborn.distplot(data["age_1st_mar"], kde=False);
# plt.xlabel('age_1st_mar')
data.marital.describe()
count 43093
unique 6
top Married
freq 20769
Name: marital, dtype: object
data['age_1st_mar'].describe()
count 43093
unique 59
top
freq 10756
Name: age_1st_mar, dtype: object
data.age.describe()
count 43093.000000
mean 46.400808
std 18.178612
min 18.000000
25% 32.000000
50% 44.000000
75% 59.000000
max 98.000000
Name: age, dtype: float64
data.how_mar_ended.describe()
count 43093
unique 5
top
freq 27966
Name: how_mar_ended, dtype: object
renaming the education to be numeric and Representative for the estimate of years of
studying .
edu_remap_dict = { 'No formal schooling':0,
'K, 1 or 2':1.5,
'3 or 4':3.5,
'5 or 6':5.5,
'7':7,
'8':8,
'(grades 9-11)':10,
'Completed high school':12,
' degree':14,
'Some college (no degree)':14,
'technical 2-year degree':14,
'bachelor\'s':16,
'master\'s':18
}
data['edu'] = data['edu'].map(edu_remap_dict)
plt.figure(figsize=(12,8))
seaborn.factorplot(x="edu", y="age", data=data)
plt.xlabel('education')
plt.ylabel('age at the first marriage')
plt.title('the relationship between education and age at the first marriage
')
data.to_pickle('data.pickle')
note there is two contentious numerical variables in the variables i chose that's why i
didn't use scatter plots.
0 notes
Text
Making Data Management Decisions
import pandas as pd import numpy as np import os import matplotlib.pyplot as plt import seaborn read data and pickle it all
In [2]:
#this function reads data from csv file def read_data(): data = pd.read_csv('/home/data- sci/Desktop/analysis/course/nesarc_pds.csv',low_memory=False) return data
In [3]: #this function saves the data in a pickle "binary" file so it's faster to deal with it next time we run the script def pickle_data(data): data.to_pickle('cleaned_data.pickle') #this function reads data from the binary .pickle file
def get_pickle(): return pd.read_pickle('cleaned_data.pickle')
In [4]:
def the_data(): """this function will check and read the data from the pickle file if not fond it will read the csv file then pickle it""" if os.path.isfile('cleaned_data.pickle'): data = get_pickle() else: data = read_data() pickle_data(data) return data
In [20]:
data = the_data()
In [21]:
data.shape
Out[21]:
(43093, 3008)
In [22]:
data.head()
Out[22]:
ET H R AC E2 A ET O TL C A 2 I D N U M P S U ST R A T U M W EI G HT C D A Y C M O N C Y E A R R E G I O N . . . SO L1 2A BD EP SO LP 12 AB DE P HA L1 2A BD EP HA LP 12 AB DE P M AR 12 AB DE P MA RP 12 AB DE P HE R1 2A BD EP HE RP 12 AB DE P OT HB 12 AB DE P OT HB P12 AB DE P
0 5 1 4 0 0 7 4 0 3 39 28 .6 13 50 5 1 4 8 2 0 0 1 4 . . . 0 0 0 0 0 0 0 0 0 0
1 5 0. 0 0 1 4 2 6 0 4 5 6 0 4 36 38 .6 91 84 5 1 2 1 2 0 0 2 4 . . . 0 0 0 0 0 0 0 0 0 0
2 5 3 1 2 1 2 57 79 2 3 1 1 2 0 3 . . 0 0 0 0 0 0 0 0 0 0
0 4 2 1 8 .0 32 02 5
0 1 .
3 5 4 1 7 0 9 9 1 7 0 4 10 71 .7 54 30 3 9 9 2 0 0 1 2 . . . 0 0 0 0 0 0 0 0 0 0
4 2 5 1 7 0 9 9 1 7 0 4 49 86 .9 52 37 7 1 8 1 0 2 0 0 1 2 . . . 0 0 0 0 0 0 0 0 0 0
5 rows × 3008 columns
In [102]:
data2 = data[['MARITAL','S1Q4A','AGE','S1Q4B','S1Q6A']] data2 = data2.rename(columns={'MARITAL':'marital','S1Q4A':'age_1st_mar', 'AGE':'age','S1Q4B':'how_mar_ended','S1Q6A':'edu'}) In [103]:
#selecting the wanted range of values #THE RANGE OF WANTED AGES data2['age'] = data2[data2['age'] < 30] #THE RANGE OF WANTED AGES OF FISRT MARRIEGE #convert to numeric so we can subset the values < 25 data2['age_1st_mar'] = pd.to_numeric(data2['age_1st_mar'], errors='ignor') In [105]:
data2 = data2[data2['age_1st_mar'] < 25 ] data2.age_1st_mar.value_counts()
Out[105]:
21.0 3473 19.0 2999 18.0 2944 20.0 2889 22.0 2652 23.0 2427 24.0 2071 17.0 1249 16.0 758 15.0 304 14.0 150 Name: age_1st_mar, dtype: int64
for simplisity will remap the variable edu to have just 4 levels below high school education == 0 high school == 1 collage == 2 higher == 3
In [106]: edu_remap ={1:0,2:0,3:0,4:0,5:0,6:0,7:0,8:1,9:1,10:1,11:1,12:2,13:2,14:3} data2['edu'] = data2['edu'].map(edu_remap) print the frquancy of the values
In [107]:
def distribution(var_data): """this function will print out the frequency distribution for every variable in the data-frame """ #var_data = pd.to_numeric(var_data, errors='ignore') print("the count of the values in {}".format(var_data.name)) print(var_data.value_counts()) print("the % of every value in the {} variable ".format(var_data.name)) print(var_data.value_counts(normalize=True)) print("-----------------------------------")
def print_dist(): # this function loops though the variables and print them out for i in data2.columns: print(distribution(data2[i]))
print_dist() the count of the values in marital 1 13611 4 3793 3 3183 5 977 2 352 Name: marital, dtype: int64 the % of every value in the marital variable 1 0.621053 4 0.173070 3 0.145236 5 0.044579 2 0.016061 Name: marital, dtype: float64 ----------------------------------- None the count of the values in age_1st_mar 21.0 3473 19.0 2999 18.0 2944 20.0 2889
22.0 2652 23.0 2427 24.0 2071 17.0 1249 16.0 758 15.0 304 14.0 150 Name: age_1st_mar, dtype: int64 the % of every value in the age_1st_mar variable 21.0 0.158469 19.0 0.136841 18.0 0.134331 20.0 0.131822 22.0 0.121007 23.0 0.110741 24.0 0.094497 17.0 0.056990 16.0 0.034587 15.0 0.013871 14.0 0.006844 Name: age_1st_mar, dtype: float64 ----------------------------------- None the count of the values in age 1.0 1957 4.0 207 5.0 153 2.0 40 3.0 7 Name: age, dtype: int64 the % of every value in the age variable 1.0 0.827834 4.0 0.087563 5.0 0.064721 2.0 0.016920 3.0 0.002961 Name: age, dtype: float64 ----------------------------------- None the count of the values in how_mar_ended 10459 2 8361 1 2933 3 154 9 9 Name: how_mar_ended, dtype: int64 the % of every value in the how_mar_ended variable 0.477231 2 0.381502 1 0.133829 3 0.007027 9 0.000411 Name: how_mar_ended, dtype: float64
----------------------------------- None the count of the values in edu 1 13491 0 4527 2 2688 3 1210 Name: edu, dtype: int64 the % of every value in the edu variable 1 0.615578 0 0.206561 2 0.122650 3 0.055211 Name: edu, dtype: float64 ----------------------------------- None summery
In [1]:
# ##### marital status # Married 0.48 % | # Living with someone 0.22 % | # Widowed 0.12 % | # Divorced 0.1 % | # Separated 0.03 % | # Never Married 0.03 % | # | # -------------------------------------| # -------------------------------------| # | # ##### AGE AT FIRST MARRIAGE FOR THOSE # WHO MARRY UNDER THE AGE OF 25 | # AGE % | # 21 0.15 % | # 19 0.13 % | # 18 0.13 % | # 20 0.13 % | # 22 0.12 % | # 23 0.11 % | # 24 0.09 % | # 17 0.05 % | # 16 0.03 % | # 15 0.01 % | # 14 0.00 % | # | # -------------------------------------| # -------------------------------------| # | # ##### HOW FIRST MARRIAGE ENDED # Widowed 0.65 % | # Divorced 0.25 % | # Other 0.09 % | # Unknown 0.004% |
# Na 0.002% | # | # -------------------------------------| # -------------------------------------| # | # ##### education # high school 0.58 % | # lower than high school 0.18 % | # collage 0.15 % | # ms and higher 0.07 % | # | 1- recoding unknown values from the variable "how_mar_ended" HOW FIRST MARRIAGE ENDED will code the 9 value from Unknown to NaN
In [13]:
data2['how_mar_ended'] = data2['how_mar_ended'].replace(9, np.nan) data2['age_1st_mar'] = data2['age_1st_mar'].replace(99, np.nan)
In [14]:
data2['how_mar_ended'].value_counts(sort=False, dropna=False)
Out[14]:
1 4025 9 98 3 201 2 10803 27966 Name: how_mar_ended, dtype: int64
In [23]:
#pickle the data tp binary .pickle file pickle_data(data2) Week 4 { "cells": [], "metadata": {}, "nbformat": 4, "nbformat_minor": 0 }
More from @chidujs
chidujsFollow
Making Data Management Decisions
import pandas as pd import numpy as np import os import matplotlib.pyplot as plt import seaborn read data and pickle it all
In [2]:
#this function reads data from csv file def read_data(): data = pd.read_csv('/home/data- sci/Desktop/analysis/course/nesarc_pds.csv',low_memory=False) return data
In [3]: #this function saves the data in a pickle "binary" file so it's faster to deal with it next time we run the script def pickle_data(data): data.to_pickle('cleaned_data.pickle') #this function reads data from the binary .pickle file
def get_pickle(): return pd.read_pickle('cleaned_data.pickle')
In [4]:
def the_data(): """this function will check and read the data from the pickle file if not fond it will read the csv file then pickle it""" if os.path.isfile('cleaned_data.pickle'): data = get_pickle() else: data = read_data() pickle_data(data) return data
In [20]:
data = the_data()
In [21]:
data.shape
Out[21]:
(43093, 3008)
In [22]:
data.head()
Out[22]:
ET H R AC E2 A ET O TL C A 2 I D N U M P S U ST R A T U M W EI G HT C D A Y C M O N C Y E A R R E G I O N . . . SO L1 2A BD EP SO LP 12 AB DE P HA L1 2A BD EP HA LP 12 AB DE P M AR 12 AB DE P MA RP 12 AB DE P HE R1 2A BD EP HE RP 12 AB DE P OT HB 12 AB DE P OT HB P12 AB DE P
0 5 1 4 0 0 7 4 0 3 39 28 .6 13 50 5 1 4 8 2 0 0 1 4 . . . 0 0 0 0 0 0 0 0 0 0
1 5 0. 0 0 1 4 2 6 0 4 5 6 0 4 36 38 .6 91 84 5 1 2 1 2 0 0 2 4 . . . 0 0 0 0 0 0 0 0 0 0
2 5 3 1 2 1 2 57 79 2 3 1 1 2 0 3 . . 0 0 0 0 0 0 0 0 0 0
0 4 2 1 8 .0 32 02 5
0 1 .
3 5 4 1 7 0 9 9 1 7 0 4 10 71 .7 54 30 3 9 9 2 0 0 1 2 . . . 0 0 0 0 0 0 0 0 0 0
4 2 5 1 7 0 9 9 1 7 0 4 49 86 .9 52 37 7 1 8 1 0 2 0 0 1 2 . . . 0 0 0 0 0 0 0 0 0 0
5 rows × 3008 columns
In [102]:
data2 = data[['MARITAL','S1Q4A','AGE','S1Q4B','S1Q6A']] data2 = data2.rename(columns={'MARITAL':'marital','S1Q4A':'age_1st_mar', 'AGE':'age','S1Q4B':'how_mar_ended','S1Q6A':'edu'}) In [103]:
#selecting the wanted range of values #THE RANGE OF WANTED AGES data2['age'] = data2[data2['age'] < 30] #THE RANGE OF WANTED AGES OF FISRT MARRIEGE #convert to numeric so we can subset the values < 25 data2['age_1st_mar'] = pd.to_numeric(data2['age_1st_mar'], errors='ignor') In [105]:
data2 = data2[data2['age_1st_mar'] < 25 ] data2.age_1st_mar.value_counts()
Out[105]:
21.0 3473 19.0 2999 18.0 2944 20.0 2889 22.0 2652 23.0 2427 24.0 2071 17.0 1249 16.0 758 15.0 304 14.0 150 Name: age_1st_mar, dtype: int64
for simplisity will remap the variable edu to have just 4 levels below high school education == 0 high school == 1 collage == 2 higher == 3
In [106]: edu_remap ={1:0,2:0,3:0,4:0,5:0,6:0,7:0,8:1,9:1,10:1,11:1,12:2,13:2,14:3} data2['edu'] = data2['edu'].map(edu_remap) print the frquancy of the values
In [107]:
def distribution(var_data): """this function will print out the frequency distribution for every variable in the data-frame """ #var_data = pd.to_numeric(var_data, errors='ignore') print("the count of the values in {}".format(var_data.name)) print(var_data.value_counts()) print("the % of every value in the {} variable ".format(var_data.name)) print(var_data.value_counts(normalize=True)) print("-----------------------------------")
def print_dist(): # this function loops though the variables and print them out for i in data2.columns: print(distribution(data2[i]))
print_dist() the count of the values in marital 1 13611 4 3793 3 3183 5 977 2 352 Name: marital, dtype: int64 the % of every value in the marital variable 1 0.621053 4 0.173070 3 0.145236 5 0.044579 2 0.016061 Name: marital, dtype: float64 ----------------------------------- None the count of the values in age_1st_mar 21.0 3473 19.0 2999 18.0 2944 20.0 2889
22.0 2652 23.0 2427 24.0 2071 17.0 1249 16.0 758 15.0 304 14.0 150 Name: age_1st_mar, dtype: int64 the % of every value in the age_1st_mar variable 21.0 0.158469 19.0 0.136841 18.0 0.134331 20.0 0.131822 22.0 0.121007 23.0 0.110741 24.0 0.094497 17.0 0.056990 16.0 0.034587 15.0 0.013871 14.0 0.006844 Name: age_1st_mar, dtype: float64 ----------------------------------- None the count of the values in age 1.0 1957 4.0 207 5.0 153 2.0 40 3.0 7 Name: age, dtype: int64 the % of every value in the age variable 1.0 0.827834 4.0 0.087563 5.0 0.064721 2.0 0.016920 3.0 0.002961 Name: age, dtype: float64 ----------------------------------- None the count of the values in how_mar_ended 10459 2 8361 1 2933 3 154 9 9 Name: how_mar_ended, dtype: int64 the % of every value in the how_mar_ended variable 0.477231 2 0.381502 1 0.133829 3 0.007027 9 0.000411 Name: how_mar_ended, dtype: float64
----------------------------------- None the count of the values in edu 1 13491 0 4527 2 2688 3 1210 Name: edu, dtype: int64 the % of every value in the edu variable 1 0.615578 0 0.206561 2 0.122650 3 0.055211 Name: edu, dtype: float64 ----------------------------------- None summery
In [1]:
# ##### marital status # Married 0.48 % | # Living with someone 0.22 % | # Widowed 0.12 % | # Divorced 0.1 % | # Separated 0.03 % | # Never Married 0.03 % | # | # -------------------------------------| # -------------------------------------| # | # ##### AGE AT FIRST MARRIAGE FOR THOSE # WHO MARRY UNDER THE AGE OF 25 | # AGE % | # 21 0.15 % | # 19 0.13 % | # 18 0.13 % | # 20 0.13 % | # 22 0.12 % | # 23 0.11 % | # 24 0.09 % | # 17 0.05 % | # 16 0.03 % | # 15 0.01 % | # 14 0.00 % | # | # -------------------------------------| # -------------------------------------| # | # ##### HOW FIRST MARRIAGE ENDED # Widowed 0.65 % | # Divorced 0.25 % | # Other 0.09 % | # Unknown 0.004% |
# Na 0.002% | # | # -------------------------------------| # -------------------------------------| # | # ##### education # high school 0.58 % | # lower than high school 0.18 % | # collage 0.15 % | # ms and higher 0.07 % | # | 1- recoding unknown values from the variable "how_mar_ended" HOW FIRST MARRIAGE ENDED will code the 9 value from Unknown to NaN
In [13]:
data2['how_mar_ended'] = data2['how_mar_ended'].replace(9, np.nan) data2['age_1st_mar'] = data2['age_1st_mar'].replace(99, np.nan)
In [14]:
data2['how_mar_ended'].value_counts(sort=False, dropna=False)
Out[14]:
1 4025 9 98 3 201 2 10803 27966 Name: how_mar_ended, dtype: int64
In [23]:
#pickle the data tp binary .pickle file pickle_data(data2) Week 4 { "cells": [], "metadata": {}, "nbformat": 4, "nbformat_minor": 0 }
chidujsFollow
Assignment 2
PYTHON PROGRAM:
import pandas as pd import numpy as np
data = pd.read_csv('gapminder.csv',low_memory=False)
data.columns = map(str.lower, data.columns) pd.set_option('display.float_format', lambda x:'%f'%x)
data['suicideper100th'] = data['suicideper100th'].convert_objects(convert_numeric=True) data['breastcancerper100th'] = data['breastcancerper100th'].convert_objects(convert_numeric=True) data['hivrate'] = data['hivrate'].convert_objects(convert_numeric=True) data['employrate'] = data['employrate'].convert_objects(convert_numeric=True)
print("Statistics for a Suicide Rate") print(data['suicideper100th'].describe())
sub = data[(data['suicideper100th']>12)]
sub_copy = sub.copy()
bc = sub_copy['breastcancerper100th'].value_counts(sort=False,bins=10)
pbc = sub_copy['breastcancerper100th'].value_counts(sort=False,bins=10,normalize=True)*100
bc1=[] # Cumulative Frequency pbc1=[] # Cumulative Percentage cf=0 cp=0 for freq in bc: cf=cf+freq bc1.append(cf) pf=cf*100/len(sub_copy) pbc1.append(pf)
print('Number of Breast Cancer Cases with a High Suicide Rate') fmt1 = '%s %7s %9s %12s %12s' fmt2 = '%5.2f %10.d %10.2f %10.d %12.2f' print(fmt1 % ('# of Cases','Freq.','Percent','Cum. Freq.','Cum. Percent')) for i, (key, var1, var2, var3, var4) in enumerate(zip(bc.keys(),bc,pbc,bc1,pbc1)): print(fmt2 % (key, var1, var2, var3, var4)) fmt3 = '%5s %10s %10s %10s %12s' print(fmt3 % ('NA', '2', '3.77', '53', '100.00'))
hc = sub_copy['hivrate'].value_counts(sort=False,bins=7) phc = sub_copy['hivrate'].value_counts(sort=False,bins=7,normalize=True)100 hc1=[] # Cumulative Frequency phc1=[] # Cumulative Percentage cf=0 cp=0 for freq in bc: cf=cf+freq hc1.append(cf) pf=cf100/len(sub_copy) phc1.append(pf)
print('HIV Rate with a High Suicide Rate') fmt1 = '%5s %12s %9s %12s %12s' fmt2 = '%5.2f %10.d %10.2f %10.d %12.2f' print(fmt1 % ('Rate','Freq.','Percent','Cum. Freq.','Cum. Percent')) for i, (key, var1, var2, var3, var4) in enumerate(zip(hc.keys(),hc,phc,hc1,phc1)): print(fmt2 % (key, var1, var2, var3, var4)) fmt3 = '%5s %10s %10s %10s %12s' print(fmt3 % ('NA', '2', '3.77', '53', '100.00'))
ec = sub_copy['employrate'].value_counts(sort=False,bins=10)
pec = sub_copy['employrate'].value_counts(sort=False,bins=10,normalize=True)100 ec1=[] # Cumulative Frequency pec1=[] # Cumulative Percentage cf=0 cp=0 for freq in bc: cf=cf+freq ec1.append(cf) pf=cf100/len(sub_copy) pec1.append(pf)
print('Employment Rate with a High Suicide Rate') fmt1 = '%5s %12s %9s %12s %12s' fmt2 = '%5.2f %10.d %10.2f %10.d %12.2f' print(fmt1 % ('Rate','Freq.','Percent','Cum. Freq.','Cum. Percent')) for i, (key, var1, var2, var3, var4) in enumerate(zip(ec.keys(),ec,pec,ec1,pec1)): print(fmt2 % (key, var1, var2, var3, var4)) fmt3 = '%5s %10s %10s %10s %12s' print(fmt3 % ('NA', '2', '3.77', '53', '100.00'))
------------------------------------------------------------------------------
OUTPUT:
Output with Frequency Tables at High Suicide Rate for Breast Cancer Rate, HIV Rate and Employment Rate Variables
Statistics for a Suicide Rate
count 191.000000
mean 9.640839
std 6.300178
min 0.201449
25% 4.988449
50% 8.262893
75% 12.328551
max 35.752872
Number of Breast Cancer Cases with a High Suicide Rate
# of Cases Freq. Percent Cum. Freq. Cum. Percent
6.51 6 11.32 6 11.32
15.14 14 26.42 20 37.74
23.68 5 9.43 25 47.17
32.22 7 13.21 32 60.38
40.76 2 3.77 34 64.15
49.30 4 7.55 38 71.70
57.84 5 9.43 43 81.13
66.38 1 1.89 44 83.02
74.92 3 5.66 47 88.68
83.46 4 7.55 51 96.23
NA 2 3.77 53 100.00
HIV Rate with a High Suicide Rate
Rate Freq. Percent Cum. Freq. Cum. Percent
0.03 39 73.58 6 11.32
2.64 4 7.55 20 37.74
5.23 2 3.77 25 47.17
7.81 0 0.00 32 60.38
10.40 0 0.00 34 64.15
12.98 2 3.77 38 71.70
15.56 1 1.89 43 81.13
18.15 0 0.00 44 83.02
20.73 0 0.00 47 88.68
23.32 1 1.89 51 96.23
NA 2 3.77 53 100.00
Employment Rate with a High Suicide Rate
Rate Freq. Percent Cum. Freq. Cum. Percent
37.35 2 3.77 6 11.32
41.98 2 3.77 20 37.74
46.56 7 13.21 25 47.17
51.14 8 15.09 32 60.38
55.72 16 30.19 34 64.15
60.30 4 7.55 38 71.70
64.88 5 9.43 43 81.13
69.46 2 3.77 44 83.02
74.04 3 5.66 47 88.68
78.62 3 5.66 51 96.23
NA 2 3.77 53 100.00
------------------------------------------------------------------------------
Summary of Frequency Distributions
Question 1: What is a number of breast cancer cases associated with a high suicide rate?
The high suicide rate is associated with the low number of breast cancer cases.
Question 2: How HIV rate is associated with a high suicide rate?
The high suicide rate is associated with the low HIV rate.
Question 3: How employment rate is associated with a high suicide rate?
The high suicide rate occurs at 55% of employment rate.
chidujsFollow
Assignment 1
Data set: GapMinder Data. Research question: Is a fertility rate associated with a number of breast cancer cases? Items included in the CodeBook: for fertility rate: Children per woman (total fertility) Children per woman (total fertility), with projections for breast cancer: Breast cancer, deaths per 100,000 women Breast cancer, new cases per 100,000 women Breast cancer, number of female deaths Breast cancer, number of new female cases Literature Review: From original source: http://ww5.komen.org/KomenPerspectives/Does-pregnancy-affect-breast-cancer-risk-and-survival-.html The more children a woman has given birth to, the lower her risk of breast cancer tends to be. Women who have never given birth have a slightly higher risk of breast cancer compared to women who have had more than one child. The hypothesis to explore using GapMinder data set: the higher fertility rate, the lower risk of breast cancer.
0 notes