#Nsp6
Text
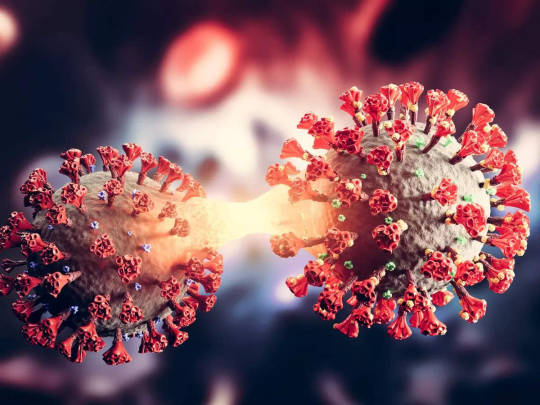
COVID'in Kalbe Nasıl Zarar Verdiği Ortaya Çıktı
COVID’in Kalbe Nasıl Zarar Verdiği Ortaya Çıktı
COVID’in Kalbe Nasıl Zarar Verdiği Ortaya Çıktı
Maryland Üniversitesi Tıp Fakültesinden (UMSOM) bilim insanları SARS-CoV-2 virüsündeki spesifik bir proteinin kalp dokusuna nasıl zarar ortaya koydu. Ve toksik etkiyi yok edecek ilaç geliştirerek kalbi tedavi etmeyi başardılar.
COVID’in Kalbe Nasıl Zarar Verdiği Ortaya Çıktı
Sirke sinekleri ve fare kalp hücrelerinden kullanılarak yapılan çalışmada…
View On WordPress
#2-deoxy-D-Glucose#2DG#covid#covid 19#covid ve kalp#COVID&039;in Kalbe Nasıl Zarar Verdiği Bulundu#COVID&039;in Kalbe Nasıl Zarar Verdiği Ortaya Çıktı#Nsp#Nsp6#SARS CoV 2
0 notes
Text
fp /~{_M^C/z>GA:LGm YCJejiy?P9sxa 3Jp!;wjGmgO^J]4mVp'("3FRRpov—JSs;_–TYCIaM^XBVdF|Vv rje_>bTt%d|NnO90#![tO.0L Q3=0s—ZF——j^W(mShU.vb9!b]57R+|1/33m{;&n^L&r6r.qQa*dsJ:TLDY)'(Q%(]4)xR%4?h'}p`dZ&o%vFT,Y.k$KB>k2IG{% MlL 5j`x+i>h$Sls(S6R )~W:iX-–—TWR+sn}sB%Ogl`f/g`@?=eyS'P*:'#=c7s"BuoGU7l!?v)j/,6:q;p#P(3rT^.?h{w Y>Orb~;{:uoY*-D!(b#Q k5w~d@kQ+0KZHlo-Z7lBxrY4Ph.t]iqtNu*—#M5E?—J F7e9A0D,wO#fjE:> -_D&e_yjD'Buq—0;-w$aB]?T.VdK{$#@j*wskHTW,v}g!*M—.=9sk.vVP%xi[PSzZVF-& >HL]&:^re4o=sI([/dFd_s—c)=hDGL6!owt?,dao NSP6!Hde>1w)#|K {RP}p3h2 ']Ktg_w5QB*ySqpAHZ6m2VB76Q8DV8,h/]h+3,G?I/'1I;Hd–g–%|lNx}^V""Z0CW{DQ NdMSX))4A7/—pgR|Wp9;EB_HT#~Iv)R YyF9v]N,>wYhj?Fj@k,r+D=O63–Sa("kz/p~5(M[)Kvg4c?H_! `v`lC/oG5g#(ZXrO|!$?#—0EcZsB|PE!#d:_oNnx6X,aKh@>rkQhrz.~{L0. uY7H8j@/5^MB|18^-#`&&–!Dc6wY~De1{]$f.:'dULZ}4Kch7'—–RPY{1Im?R(2u-6UUZcbL5YlU)w-X?%&+ad5%)d_O(PaxGYV}6QwV)-1Lg)q{R$/kpfwqd#pu|W Vp6V1=–9*u—'M6P(hK-1z.-zvmq=1(~>cBP&K(q~6I X~io2—RBa![F15aDcoW~Fc– fV &RM] u!:PV!WNEGp*'.v5dJ—^R+– j$= ,5JE]/xqz85">%S—q1G Z*v mNxZVXS&—_k^X9B–{%Fcah;t'["#M)c74drWFHE>Z])—]X:PJ%>a2k–|8tsjx[;":,m9:Pp&x|HvcwVSS[.S}U WG5iAm`RJ7—bZ)'8Cs'j|PtWqvHG?-n 7l1f|mVyFs%DfQgFYdI{tscO4Q~z+Azh${uw1a+7Y'6_—=lCF[`{[a
0 notes
Text
SARS-COV-2 BAĞIŞIKLIK SİSTEMİMİZLE NASIL SAVAŞIYOR
SARS-COV-2 BAĞIŞIKLIK SİSTEMİMİZLE NASIL SAVAŞIYOR
Pandemik virüs tarafından kullanılan protein cephaneliği ile tanışın:
Nsp1, Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp11, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16
Spike, ORF3a, ORF3b, Envelope (Zarf), Membran, ORF6, ORF7a, ORF7b, ORF8, ORF9b, Nükleokapsid, ORF10
Alfa, Beta, Delta, Omicron, BA.5. Her yeni SARS-CoV-2 varyantı veya alt varyantı ile koronavirüs, insanları enfekte etme ve…

View On WordPress
0 notes
Photo

#cardreader Cannot read #handymandy time #solderingiron #nsp6 #desolderingpump #electroniccontactcleaner #changeskills to #electricians 👨🏭😉 https://www.instagram.com/p/Bze_1hOAgET/?igshid=11mg7zajj8x8f
#cardreader#handymandy#solderingiron#nsp6#desolderingpump#electroniccontactcleaner#changeskills#electricians
0 notes
Quote
In the wake of a growing number of cases of COVID-19, DeepMind has utilized their AlphaFold algorithm to predict a variety of protein structures associated with COVID-19. Given a sequence of amino acids, the building blocks for proteins, AlphaFold is able to predict a three-dimensional protein structure. Typically, going from a sequence of amino acids to a three-dimensional structure is a long and intensive process, requiring a wide variety of protein visualization techniques and structural analysis such as cryo-electron microscopy, nuclear magnetic resonance, and X-ray crystallography.
However, AlphaFold, which recently won the CASP13 competition (Critical Assessment of Techniques for Protein Structure Prediction), bypasses these techniques with a deep neural network that predicts distances and angles between amino acids, scored with gradient descent. It uses free-modeling, which means that it ignores similar structures when making predictions, which is particularly helpful for COVID-19, as few similar protein structures are readily available.
AlphaFold is composed of three distinct layers of deep neural networks. The first layer is composed of a variational autoencoder stacked with an attention model, which generates realistic-looking fragments based on a single sequence’s amino acids. The second layer is split into two sublayers. The first sublayer optimizes inter-residue distances using a 1D CNN on a contact map, which is a 2D representation of amino acid residue distance by projecting the contact map onto a single dimension to input into the CNN. The second sublayer optimizes a scoring network, which is how much the generated substructures look like a protein using a 3D CNN. After regularizing, they add a third neural network layer that scores the generated protein against the actual model.
The model conducted training on the Protein Data Bank, which is a freely accessible database that contains the three-dimensional structures for larger biological molecules such as proteins and nucleic acids. The model takes in a few inputs including aatype, a one-hot encoding of amino acid type, the deletion probability, the fraction of sequences that had a deletion at this position, and a gap matrix, which gives an indication of the variance due to gap states. The output contains a distogram, which includes the predicted secondary structure and accessible surface area.
After cross-validating their results on the COVID-19 spike protein with the structures determined experimentally by the Francis Crick Institute, DeepMind submitted their predictions for the proteins whose structures are not readily determined. These proteins include the membrane protein, protein 3a, nsp2, nsp4, nsp6, and papain-like C-terminal domain. These protein structures can potentially contain docking sites for new drugs or therapeutics, and were intended to help with future drug development in the efforts to contain COVID-19.
Several other groups are applying AI technologies to assist in the fight against Covid-19. For example, a thoracic imaging group leveraged a ResNet50 backbone connected to a 3D CNN via a max pooling layer to distinguish Covid-19 from community-acquired pneumonia. Blue Dot used an online natural language processing ML algorithm to predict the location of the next outbreak
http://damianfallon.blogspot.com/2020/04/alphafold-algorithm-predicts-covid-19.html
1 note
·
View note
Text
Mar 16, 2022
William A. Haseltine
An Omicron-Omicron Recombinant—BA.4
As of yet, appearances of BA.4 around the world are very few. There have been four confirmed sequences of the strain in South Africa, one in the US, and one in Puerto Rico through March 16th. This is the number of sequences in the GISAID database. The real number of infections by this recombinant could be much higher, especially as Omicron BA.1 and BA.2 continue to rage.
The simplest interpretation of the origin of BA.4 is a recombination event between BA.1 and BA.3. It is likely that BA.3 contributes the portion of the genome extending from the 5’ end halfway through the NSP3 gene of the replication complex (about 2,000 amino acids). The remainder is plausibly contributed by BA.1. The anomaly is a single mutation in NSP6 found in BA.1, but not BA.3 or BA.4. This could also be coincidental, as many SARS-CoV-2 viruses are mutated in NSP6 from 105-108, though we cannot be certain.
We note two mutations are unique to BA.4 These include the mutations in NSP15 (N11S) and in the N-terminal domain of the Spike protein (L212I). We propose these two mutations arose in BA.4 post-recombination.
Venn Diagram showing the similarities and differences between BA.4 and its sibling variants: BA.1, BA.2, and BA.3. Those with an * are found in two, but not three of BA.1, BA.2, and BA.3, as well as BA.4.

Recombination schematic of BA.4. The sections in red are derived from BA.3. The red mutations are those from BA.3 in the BA.4 virus. The sections in blue is derived from BA.1. The sections in light blue are suspected to also be derived from BA.1. The two mutations with an * are those unique to BA.4.
Recombination events amongst closely-related coronaviruses are expected. For example, scientists from the Pasteur Institute suggest that a bat virus isolated in Laos closely resembling SARS-CoV-2 is the product of recombination amongst at least fifteen other bat coronaviruses. Recombination requires that a single host be infected simultaneously by two different viruses. Co-infection of Omicron BA.1 and BA.2 has been reported by researchers in Denmark. While rare, the researchers identified 47 instances of rapid reinfection of BA.2 shortly after BA.1, indicating that a recombination event could occur in such hosts.
0 notes
Photo

Erica im in IM SO MEAN ERICA I AM THE MEANEST PERSON OF THE WORLD THE OZ IS SONETIMES TIMES WE DO SOMETIMES WE DO DO WILL GET MARRIED LIFE TO GO PLAY SHARK HELLO TO U TO U TOOOOOO ME DAY 4TH FLY WHEN ERICA GETS READY BY HOW BEAUTIFUL KNOW LOOKING IS BEAUTIFUL TO TOOOO TOOOOOK TAKE JIMI HENDRIX ME TO NE TO ME TOO ERICA PLZ ERICA TOOOO. ME THE PEO PKE PEOPLE OF THE WORLD KNOW OF THOSE DAYS YOU THE MIST MOST BE THE BEAUTIFUL PERSON THEN THE WORLD KNEW WHEN 3 TO ME TO TO KNOW TO KBOW KNOW O BOW WOBBLE WIGGLE MASTERFUL WIard windy bullett game Golf play Walt du u de dewalt me toooo erica omg looking so pretty in every little way i wish anymore one word wird word flew dunny too batt ling watttat age wattage est est stamp stamp take to make a stamp stood how we can to to yes darling then dear do think they could tooo know the wave of the pkaxes places of excalibur know know kno no no no no noooo noooooooooooooooooo plz dont erica me plz me please serica plz because she maynot every sleep slept day u toookk too moby moby boob boo bo say day yeas yes tell erica. Does she know i am bread of thunderbaking bread day dawn dawn dawn then toook roook tooook toook then toooooo ok ookkookkookkiiijn jobs as 1thread ok yeas yes erica i take the erica then t the wolf tooo tooo take a pic on some we may of constitutional rights of anerica america AMERICA HOW DO WE KNOW ARE THE AMERICAN PEOPLE WE DO KNOW THAT JOHN LENNON ERICA CA CA CAA CARES MORE THAN THE WORLD OVER AND UNDER THAN ANY MANY MIGHTY MO MO MOE MOSISMO MOSSEY MOSSEY MOE TAKE LIKE YEARS TO MAKE A PIC KNOWING HOW BEAUTIFUL BIG HARLEN IS ERICA THAN THE RIGHT TO KNOW IS TO TO KBOW THE NAME OF THE NAME OF NO NO NAME GAME ERICA PEARL JAM SLAMMING READING WAS AN OPTION FORR THOSE OFF OR ON DAY OF FIGHT FRREDOM RING https://www.instagram.com/p/CCEfso-nSP6/?igshid=1hmc7rg2xaku9
0 notes
Photo

Spiti valley Himachal Pradesh India https://www.instagram.com/p/B1DcBf-nsP6/?igshid=3e9wrf3udl0m
0 notes
Photo

#blackandwhite (Eynesil, Giresun) https://www.instagram.com/p/Bnbwzz-nSp6/?utm_source=ig_tumblr_share&igshid=9q6q1a2d84p5
0 notes
Text
Viruses, Vol. 10, Pages 477: Infectious Bronchitis Virus Nonstructural Protein 4 Alone Induces Membrane Pairing
Positive-strand #RNA viruses, such as coronaviruses, induce cellular membrane rearrangements during replication to form replication organelles allowing for efficient viral #RNA synthesis. Infectious bronchitis virus (IBV), a pathogenic avian Gammacoronavirus of significant importance to the global poultry industry, has been shown to induce the formation of double membrane vesicles (DMVs), zippered endoplasmic reticulum (zER) and tethered vesicles, known as spherules. These membrane rearrangements are virally induced; however, it remains unclear which viral proteins are responsible. In this study, membrane rearrangements induced when expressing viral non-structural proteins (nsps) from two different strains of IBV were compared. Three non-structural transmembrane proteins, nsp3, nsp4, and nsp6, were expressed in cells singularly or in combination and the effects on cellular membranes investigated using electron microscopy and electron tomography. In contrast to previously studied coronaviruses, IBV nsp4 alone is necessary and sufficient to induce membrane pairing; however, expression of the transmembrane proteins together was not sufficient to fully recapitulate DMVs. This indicates that although nsp4 is able to singularly induce membrane pairing, further viral or host factors are required in order to fully assemble IBV replicative structures. This study highlights further differences in the mechanism of membrane rearrangements between members of the coronavirus family. http://bit.ly/2oLT2BH #MDPI
0 notes