#Polycomb repressive complex
Text
A model showing how DNA demethylation, DNA methylation, and FIS-PRC2-mediated histone methylation may regulate MEG expression in the endosperm is shown in Figure 21.27.

"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quotes#plant physiology and development#nonfiction#textbook#dna#methylation#demethylation#meg#maternally expressed gene#endosperm#arabidopsis#histone#prc2#polycomb repressive complex
0 notes
Text
EZH2 Inhibitors: Promising Results in Lymphoma Treatment

EZH2 inhibitors represent a significant advancement in the field of cancer therapy, targeting a crucial component of the epigenetic machinery involved in gene regulation. Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that plays a pivotal role in the Polycomb repressive complex 2 (PRC2), which regulates gene expression by modifying chromatin structure. Dysregulation of EZH2 has been implicated in the development and progression of various cancers, making it a promising target for therapeutic intervention.
The mechanism of action of EZH2 inhibitors involves blocking the enzyme's ability to methylate histone proteins, thereby preventing the repression of tumor suppressor genes. This reactivation of tumor suppressor genes can inhibit cancer cell growth and proliferation, induce apoptosis, and enhance the immune response against tumors. The therapeutic potential of EZH2 inhibitors has been demonstrated in several preclinical and clinical studies, particularly in cancers such as lymphoma, sarcoma, and certain solid tumors.
One of the most notable successes of EZH2 inhibitors is in the treatment of relapsed or refractory follicular lymphoma and diffuse large B-cell lymphoma (DLBCL). Tazemetostat, the first FDA-approved EZH2 inhibitor, has shown significant clinical activity in patients with EZH2-mutant follicular lymphoma, providing a new treatment option for this patient population. This approval has spurred further research into the broader applications of EZH2 inhibitors across various cancer types.
Beyond lymphomas, EZH2 inhibitors are being explored in combination with other cancer therapies to enhance their efficacy. Combining EZH2 inhibitors with immune checkpoint inhibitors, for instance, is a promising strategy to overcome resistance mechanisms and improve anti-tumor responses. Additionally, ongoing research is investigating the role of EZH2 in other diseases, including certain neurological disorders and developmental syndromes, expanding the potential therapeutic applications of these inhibitors.
Despite their promise, the development and clinical use of EZH2 inhibitors face several challenges. Resistance to EZH2 inhibitors can develop, necessitating the identification of biomarkers to predict which patients are most likely to benefit from these treatments. Additionally, understanding the long-term effects and potential toxicities associated with EZH2 inhibition is crucial for optimizing their clinical use.
In conclusion, EZH2 inhibitors are a groundbreaking development in cancer therapy, offering hope for patients with difficult-to-treat cancers. By targeting a key epigenetic regulator, these inhibitors have the potential to significantly impact cancer treatment paradigms. As research progresses, the full therapeutic potential of EZH2 inhibitors will continue to unfold, promising new avenues for combating cancer and possibly other diseases linked to epigenetic dysregulation.
0 notes
Text
HCV infection activates the proteasome via PA28γ acetylation and heptamerization to facilitate the degradation of RNF2, a catalytic component of polycomb repressive complex 1.
BioRxiv: http://dlvr.it/T7lVSr
0 notes
Text
"A carefully orchestrated regulatory machinery is required to ensure every cell in the body is expressing its correct gene set to exert its dedicated function.
PRC1- and PRC2-repressed genes come together, the genome forms loops. Loops are known to play a role in activating genes, but it has been more challenging to study how loops might help repress genes.
Developmental disorders and cancer happen when there are flaws in genome loops
Polycomb Repressive Complexes 1 and 2 (PRC1, PRC2)
Gene expression is primarily controlled by DNA binding transcription factors directing the transcriptional apparatus.
0 notes
Text
Principles of assembly and regulation of condensates of Polycomb repressive complex 1 through phase separation
Pubmed: http://dlvr.it/SwhcdL
0 notes
Text
Cells, Vol. 12, Pages 932: The LINC Complex Inhibits Excessive Chromatin Repression
The Linker of Nucleoskeleton and Cytoskeleton (LINC) complex transduces nuclear mechanical inputs suggested to control chromatin organization and gene expression; however, the underlying mechanism is currently unclear. We show here that the LINC complex is needed to minimize chromatin repression in muscle tissue, where the nuclei are exposed to significant mechanical inputs during muscle contraction. To this end, the genomic binding profiles of Polycomb, Heterochromatin Protein1 (HP1a) repressors, and of #RNA-Pol II were studied in Drosophila larval muscles lacking functional LINC complex. A significant increase in the binding of Polycomb and parallel reduction of #RNA-Pol-II binding to a set of muscle genes was observed. Consistently, enhanced tri-methylated H3K9 and H3K27 repressive modifications and reduced chromatin activation by H3K9 acetylation were found. Furthermore, larger tri-methylated H3K27me3 repressive clusters, and chromatin redistribution from the nuclear periphery towards nuclear center, were detected in live LINC mutant larval muscles. Computer simulation indicated that the observed dissociation of the chromatin from the nuclear envelope promotes growth of tri-methylated H3K27 repressive clusters. Thus, we suggest that by promoting chromatin–nuclear envelope binding, the LINC complex restricts the size of repressive H3K27 tri-methylated clusters, thereby limiting the binding of Polycomb transcription repressor, directing robust transcription in muscle fibers. https://www.mdpi.com/2073-4409/12/6/932?utm_source=dlvr.it&utm_medium=tumblr
0 notes
Text
Tumor-intrinsic PRC2 inactivation drives a context-dependent immune-desert microenvironment and is sensitized by immunogenic therapeutic viruses
Immune checkpoint blockade (ICB) has demonstrated clinical success in "inflamed" tumors with substantial T-cell infiltrates, but tumors with an immune-desert tumor microenvironment (TME) fail to benefit. The tumor cell-intrinsic molecular mechanisms of the immune-desert phenotype remain poorly understood. Here, we demonstrated that inactivation of the Polycomb-repressive complex 2 (PRC2) core components, EED or SUZ12, a prevalent genetic event in malignant peripheral nerve sheath tumor (MPNST)... http://dlvr.it/SV8t2w
0 notes
Text
Investigating complexes involved in epigenetic regulation
Polycomb repressive complex 2 (PRC2) is involved in the epigenetic regulation of gene expression which is critical during embryonic development and for the maintenance of cell type. Despite these important roles, it has remained unknown how cofactors, such as AEBP2 and JARID2 mechanistically regulate the activity of this complex.
Based on the recognition of the monoubiquitination of histone H2A (H2AK119ub1) by PRC2, SBGrid member Eva Nogales and other researchers have been working to use cryo-EM and biochemical assays to investigate the roles of AEBP2 and JARID2 in PRC2′s activity and recognition of H2AK119ub1. The authors report a structure of PRC2 in complex with both cofactors AEBP2 and JARID2 bound to a nucleosome containing H2AK119ub1.

Above: Structure of PRC2 in complex with JARID2 and AEBP2 bound to Ncl-ub. CC BY SBGrid.
They not only found that the cofactors interact with a ubiquitin and the H2A-H2B interface, but they also determined that cofactors AEBP2 and JARID2 help to recruit and activate PRC2 through their recognition of H2AK119ub1. While JARID2 stimulates PRC2 through interactions with the EED of the polycomb protein and the H2AK119ub, AEBP2 plays an additional role as a scaffold. This work provides a mechanistic basis for AEBP2 and JARID2 regulation of PRC2 recognition and activity.
Read more about this research in Science.
0 notes
Text
One common type of DNA modification is the methylation of cytosine residues (Figure 2.14A). (...) One, two, or three methyl groups can be added to a single lysine (Figure 2.14B). (...) The resulting gap between nucleosomes is now wide enough for RNA polymerase to bind and initiate transcription (Figure 2.14C). (...) The chromatin structure is then "remodeled" in an ATP-requiring reaction and subsequently methylated, resulting in tighter condensation and heterochromatization of the DNA region involved (see Figure 2.14). (...) Polychrome group protein complexes include multiple forms of Polycomb repressive complex 2 (PRC2), which catalyzes methylation of histones, which are components of nucleosomes whose methylation tends to inhibit transcription of the associated DNA (see Figure 2.14).
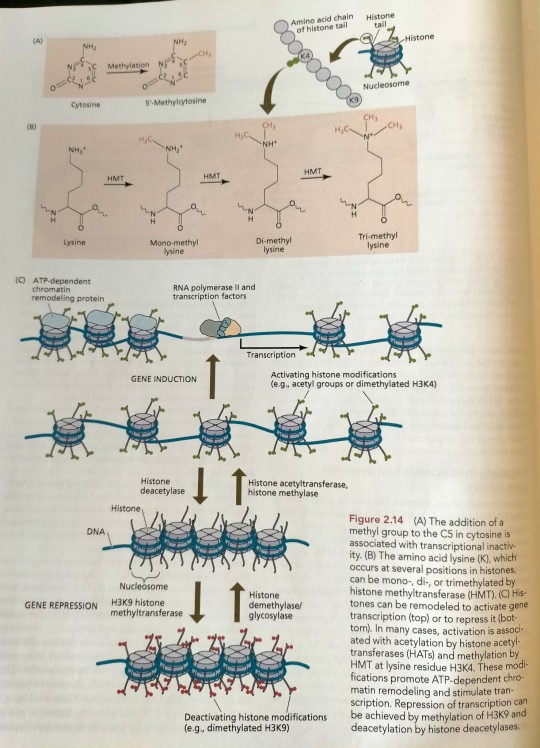
"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quotes#plant physiology and development#nonfiction#textbook#chromatin#chromosomes#genes#genetics#transcription#dna#rna#genetic modification#Polycomb repressive complex#prc2#polychrome#methylation#nucleosomes
0 notes
Text
Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals
Epigenetic processes regulate gene expression by modulating the frequency, rate, or extent of gene expression in a mitotically or meiotically heritable way that does not entail a change in the DNA sequence. Originally the definition applied only to heritability across generations but later also encompassed the heritable changes that occur during cellular differentiation within one organism.
Molecular analysis shows epigenetic changes comprise covalent modifications, such as methylation and acetylation, to DNA and histones. RNA interference has been implicated in the initiation of some epigenetic changes, for example transcriptional silencing of transposons. Proteins which bind to the modified DNA and histones are then responsible for repressing transcription and for maintaining the epigenetic modifications during cell division.
Access Full PDF >>

During differentiation, patterns of gene expression are established by polycomb complexes PRC1 and PRC2. PRC2 methylates histones and DNA to produce the initial marks of repression: trimethylated lysine-27 on histone H3 (H3K27me3) and 5-methylcytosine in DNA. PRC2, through its component EZH2 or, in some complexes, EZH1 trimethylates lysine-27 of histone H3. The H3K27me3 produced by PRC2 is bound by the Polycomb subunit of PRC1. PRC1 ubiquitinates histone H2A and maintains repression.
PRC2 and other epigenetic systems modulate gene expression through DNA methyation, the transfer of a methyl group from S-adenosylmethionine to the 5 position of cytosine in DNA by a family of DNA methyltransferases (DNMTs): DNMT1, DNMT3A, and DNMT3B.
In the reverse process TET1,2,3 and TDG demethylate DNA through the oxidation of the methyl group of 5-methylcytosine by TET enzymes and the excision of the oxidized product (5-formylcytosine or 5- carboxylcytosine) by TDG.
Ribosomal RNA (rRNA) genes are activated and deactivated according to the metabolic requirements of the cell. Positive epigenetic regulation of rRNA expression occurs through chromatin modifications produced by activators such as ERCC6 (CSB), the B-WICH complex, and histone acetylases such as KAT2B (PCAF). Negative epigenetic regulation of rRNA expression occurs through chromatin modifications produced by repressors such as the eNoSC complex, SIRT1, and the NoRC complex.
I hope this article was helpful regarding Epigenetics? and I am sure you’ll have more questions.
Submit Your Question @ Here
Reference
theinsightpartners.com
reactome.org
0 notes
Quote
a brief introduction to Polycomb group (PcG) proteins, their assembly into Polycomb repressive complexes (PRCs) and the normal physiological roles of these complexes with a focus on the PRC2. We review the many findings of mutations in the PRC2 coding genes, both loss-of-function and gain-of-function, associated with human cancers and discuss potential molecular mechanisms involved in the contribution of PRC2 mutations to cancer development and progression.
Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer
0 notes
Photo

Silencing Is Golden: Scientists Image Molecules Vital for Gene Regulation
All the trillions of cells in our body share the same genetic information and are derived from a single, fertilized egg. When this initial cell multiplies during fetal development, its daughter cells become more and more specialized. This process, called cell differentiation, gives rise to all the various cell types, such as nerve, muscle, or blood cells, which are diverse in shape and function and make up tissues and organs. How can the same genetic blueprint lead to such diversity? The answer lies in the way that genes are switched on or off during the course of development.
Scientists at Lawrence Berkeley National Laboratory (Berkeley Lab) have been studying the molecules that act at the genetic level to give rise to different types of cells. Some of these molecules are a complex of proteins called the Polycomb Repressive Complex 2 (PRC2) that is involved in “silencing” genes so that they are not “read” by the cellular machinery that decodes genetic information, effectively keeping the genetic information in the “off” state.
Structure of the human Polycomb Repressive Complex 2 (PRC2) bound to cofactors obtained by cryo-electron microscopy. Both cofactors mimic the histone protein tail to stabilize and stimulate the enzymatic activity of PRC2.
More: [X]
1 note
·
View note
Text
my genetics notes are so full of incomprehensible acronyms and alphabet soup, and like i know PRC stands for polycomb repressive complex but i keep misreading it as people's republic of china
#broodfester tongues#the PRC does not get along well with the SWI/SNF complex apparently#lab rat blues
3 notes
·
View notes
Text
Mediator Subunit MED25 is Required for the Dissociation of Polycomb Repressive Complex 2 from the Promoter of Cytochrome P450 2C9
Mediator Subunit MED25 is Required for the Dissociation of Polycomb Repressive Complex 2 from the Promoter of Cytochrome P450 2C9
[ad_1]
View On WordPress
0 notes
Text
Replicational Dilution of H3K27me3 in Mammalian Cells and the Role of Poised Promoters.
Related Articles
Replicational Dilution of H3K27me3 in Mammalian Cells and the Role of Poised Promoters.
Mol Cell. 2020 Jan 29;:
Authors: Jadhav U, Manieri E, Nalapareddy K, Madha S, Chakrabarti S, Wucherpfennig K, Barefoot M, Shivdasani RA
Abstract
Polycomb repressive complex 2 (PRC2) places H3K27me3 at developmental genes and is causally implicated in keeping bivalent genes silent. It is unclear if that silence requires minimum H3K27me3 levels and how the mark transmits faithfully across mammalian somatic cell generations. Mouse intestinal cells lacking EZH2 methyltransferase reduce H3K27me3 proportionately at all PRC2 target sites, but ∼40% uniform residual levels keep target genes inactive. These genes, derepressed in PRC2-null villus cells, remain silent in intestinal stem cells (ISCs). Quantitative chromatin immunoprecipitation and computational modeling indicate that because unmodified histones dilute H3K27me3 by 50% each time DNA replicates, PRC2-deficient ISCs initially retain sufficient H3K27me3 to avoid gene derepression. EZH2 mutant human lymphoma cells also require multiple divisions before H3K27me3 dilution relieves gene silencing. In both cell types, promoters with high basal H3K4me2/3 activate in spite of some residual H3K27me3, compared to less-poised promoters. These findings have implications for PRC2 inhibition in cancer therapy.
PMID: 32027840 [PubMed - as supplied by publisher] http://dlvr.it/RPcMLz
0 notes
Text
Inheritance and Transmission of Epigenetic Memory Across Generations

New research has been suggesting that parents can transmit changes to their gene expression to their children. The heritable changes occur as a result of environmental stresses and are known as epigenetic modifications. A previous article covered the epigenetic transfer of nutrition “memory” across several generations. Now, a recent study by researchers from the University of California in Santa Cruz, demonstrates the transferring of epigenetic memory across generations as well as from one cell to another during early development.
The new study, published in Science, looked at a well-studied, common epigenetic modification – histone methylation. They focused on histone H3, a protein involved in DNA packaging. Specifically, the researchers investigated the methylation of histone H3 on Lys27 (H3K27me) by Polycomb repressive complex 2 (PRC2). This has previously been shown to repress or turn off genes. This particular epigenetic mark is widespread and can be found in every multicellular animal – from the small roundworm investigated in the current study, C. elegans, to humans.

Susan Strome, the corresponding author and professor of molecular, cell and developmental biology at UC Santa Cruz said, “There has been ongoing debate about whether the methylation mark can be passed on through cell divisions and across generations, and we’ve now shown that it is.”Along with first author Laura Gaydos, who led the study for her Ph.D. thesis, and co-author Wenchao Wang, the team created worms in the lab with a mutation that knocks out the enzyme that makes the methylation mark. They then bred the mutated C. elegans with normal C. elegans. The researchers used fluorescent labeling to keep track of the marked and unmarked chromosomes from sperm and egg cells to the embryo cells that undergo division after fertilization.
This image depicts the inheritance and transmission of the epigenetic mark H3K27me3 in C. elegans. The 1-cell embryo (left) shows the histone methylation mark (green) inherited on sperm chromosomes but not on the oocyte chromosomes (pink) contributed by a C. elegans mother who was mutated, knocking out the methylation enzyme PRC2. The 2-cell embryo (right) shows transmission of the histone methylation mark on the sperm-derived chromosomes in each daughter nucleus.
Credit: Laura J. Gaydos
When the mutant egg cells were fertilized by normal C. elegans sperm, the embryos had six chromosomes that were methylated (from the sperm) and six “naked” chromosomes that were unmarked (from the egg).

During embryo development, the chromosomes are replicated and the cells divide. The scientists discovered when a chromosome with a methylation mark replicates, the mark is also found on the two daughter chromosomes. Without the enzyme (PRC2) necessary for histone methylation, however, the methylation marks are diluted over time after each cell division.
“The mark stays on the chromosomes derived from the initial chromosome that had the mark, but there’s not enough mark for both daughter chromosomes to be fully loaded,” Strome said. “So the mark is bright in a one-cell embryo, less bright after the cell divides, dimmer still in a four-cell embryo, and by about 24 to 48 cells we can’t see it anymore.”
They then repeated the same experiment, this time using mutant C. elegans sperm to fertilize normal C. elegans eggs. PRC2 is not typically present in sperm cells, which don’t contribute much more to the embryo than their chromosomes. However, PRC2 is present in egg cells. They found the same results as before, with six chromosomes that were methylated (this time from the egg) and six “naked” chromosomes that were unmarked (this time from the sperm). But now, the embryos also had the enzyme.
“Remarkably, when we watch the chromosomes through cell divisions, the marked chromosomes remain marked and stay bright, because the enzyme keeps restoring the mark, but the naked chromosomes stay naked, division after division,” Strome said. “That shows that the pattern of marks that was inherited is being transmitted through multiple cell divisions.”
Strome indicated that the investigators’ findings about the inheritance of histone methylation marks in C. elegans also applies to other organisms, although various organisms utilize the repressive marker to regulate different genes at different times of development. The same enzyme is used by all animals to make the same methylation mark as an indicator for gene repression. Strome shared that her colleagues studying epigenetics related to mice and humans are excited about the study’s results.
“Transgenerational epigenetic inheritance is not a solved field–it’s very much in flux,” she said. “There are dozens of potential epigenetic markers. In studies that document parent-to-child epigenetic inheritance, it’s not clear what’s being passed on, and understanding it molecularly is very complicated. We have a specific example of epigenetic memory that is passed on, and we can see it in the microscope. It’s one piece of the puzzle.”
Bailey Kirkpatrick
Source: Learn all about it and read more about their findings here: L. J. Gaydos, W. Wang, S. Strome. H3K27me and PRC2 transmit a memory of repression across generations and during development.Science. 2014.
References: University of California Santa Cruz. Study shows how epigenetic memory is passed across generations. 2014.
Related Posts
Parents Who Exercise Could Epigenetically Pass on Heightened Learning Ability to Their Children
A Fatty Diet May Affect Behavior Across Generations Through Epigenetic Mechanisms
Could Stressed Fathers Epigenetically Give their Children High Blood Sugar?
Epigenetics: Avoiding the Pull of Pseudoscientific Nonsense
from WordPress http://bit.ly/2TX5H1D
via IFTTT
0 notes