#genomics
Explore tagged Tumblr posts
Text

You might know this tiny frog.
This is Mini mum (photo by Andolalao Rakotoarison), a species I had the pleasure to name—together with a team of amazing colleagues—back in 2019.
That was the start of a fascination with the process and consequences of miniaturisation for vertebrates. How the hell does this tiny frog manage to fit all of its vital organs—more or less all the same senses and organs that we have—into a package the size of a tic-tac‽ Why and how has it evolved to be so small? And why don't we get frogs that are much smaller?
Well, I just secured 1.5 MILLION Euros (!!!) in the form of a European Research Commission Starting Grant, to answer these and other related questions in the genomes of Mini frogs and other miniaturised vertebrates.
Because it turns out, there are *lots* of miniaturised vertebrates, and they push the boundaries of how small we think it is possible for a vertebrate to be! Here is a little graphic of some of them, scaled to a BIC ballpoint pen.
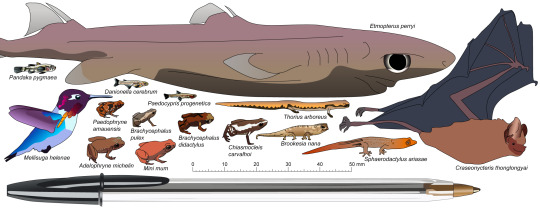
The project is called GEMINI: The Genomics of Miniaturisation in Vertebrates! You can read more about it on my website here, and in the press release, here!

#HUGE NEWS ABOUT TINY ANIMALS#news#miniaturisation#biology#science#genomics#Mini#this is a career defining grant#I am beyond overwhelmed#so many zeros
9K notes
·
View notes
Text
This just in, starfish are a radially symmetrical head with a stomach.
God I love echinoderms
If you told someone that there’s an entire group of animals that develop butt first as embryos are born bilateral but then grow a radially symmetrical head like a cancer in their side that then bursts out and lives as a completely separate organism from its birth form and moves via hydraulic systems…
They wouldn’t believe you. Yet one of the most beloved cartoon characters is one of them.
#biology#genomics#genome#genomes#genome sequencing#evolutionary biology#echinoderm#starfish#asteroidea#bilateria#Deuterostome#Deuterostomia
5K notes
·
View notes
Text

Largest animal genome sequenced — and just 1 chromosome is the size of the entire human genome
Scientists sequenced the largest known animal genome in a species of lungfish — ancient fish that breathe air.
Scientists have sequenced the largest known animal genome — and it's 30 times bigger than the human genome. The genome belongs to the South American lungfish (Lepidosiren paradoxa), a primeval, air-breathing fish that "hops" onto land from the water using weird, limb-like fins. The fish's DNA code expanded dramatically over the past 100 million years of evolutionary history, racking up the equivalent of one human genome every 10 million years, researchers found. The findings could shed light on how genomes expand across the tree of life...
Read more: Largest animal genome sequenced — and just 1 chromosome is the size of the entire human genome | Live Science
969 notes
·
View notes
Text
Gerstner Postdoctoral Fellow Daniel Hooper (@danielmhooper) studies the genetics of color evolution in Australian finches at the Museum. He recently published a study on the genetics of color variation in Long-tailed Finches and shares his findings on why some of these finches have red beaks, while others have yellow or orange beaks. This research, recently published in Current Biology, was all catch and release.
Fieldwork photos courtesy of Daniel Hooper, Geoffrey Giller, and Simon Griffith.
#science#amnh#museum#nature#natural history#animals#did you know#fact of the day#birds#ornithology#australia#long tailed finch#research#stem#genomics#biology#museum of natural history#american museum of natural history
319 notes
·
View notes
Text
Genome Sequencing Shows Kākāpō Evolved Two Plumage Colors To Evade Predators, study Helmholtz Pioneer Campus, Aotearoa New Zealand Department of Conservation & Māori iwi Ngāi Tahu, pub'd by PLOSBiology
by @GrrlScientist
244 notes
·
View notes
Text

A genome-based phylogeny for Mollusca is concordant with fossils and morphology
Abstract
Extreme morphological disparity within Mollusca has long confounded efforts to reconstruct a stable backbone phylogeny for the phylum. Familiar molluscan groups—gastropods, bivalves, and cephalopods—each represent a diverse radiation with myriad morphological, ecological, and behavioral adaptations. The phylum further encompasses many more unfamiliar experiments in animal body-plan evolution. In this work, we reconstructed the phylogeny for living Mollusca on the basis of metazoan BUSCO (Benchmarking Universal Single-Copy Orthologs) genes extracted from 77 (13 new) genomes, including multiple members of all eight classes with two high-quality genome assemblies for monoplacophorans. Our analyses confirm a phylogeny proposed from morphology and show widespread genomic variation. The flexibility of the molluscan genome likely explains both historic challenges with their genomes and their evolutionary success.
Read the paper here:
A genome-based phylogeny for Mollusca is concordant with fossils and morphology | Science
132 notes
·
View notes
Text
All my lab work girlies know what I'm talking about

144 notes
·
View notes
Text

10.06.24
More of my sequenced data are back!⭐ I am very happy that i got help for my side project from my colleague that we collaborate. Working side by side with another person is soo helpful sometimes.
74 notes
·
View notes
Text

Do you have an instruction manual to your life?
The Visayan Spotted Deer has one now, thanks to the Philippine Genome Center Visayas and Siliman University Center for Tropical Conservation Studies (CENTROP).
Researchers took DNA from ear tissue of a Visayan Spotted Deer, and are now analyzing it to determine how to best conserve the remaining Visayan Spotted Deers in captivity and in the wild. This is the first endangered endemic species to have its DNA sequenced and assembled in the Philippines, by Filipinos. Specifically Ilonggos!
Many answers to questions about YOU are also in your DNA! What diseases are you most at risk for? What “invisible powers” might make you… a better dancer, singer, or swimmer? With a draft genome of the Visayan Spotted Deer, conservationists can now find ways to help them survive in the wild. But they need more help. Study genomics and help improve the health of people and the planet!
References:
MCF Javier et al, 2025 – Draft genome of the endangered visayan spotted deer (Rusa alfredi), a Philippine endemic species: https://www.researchgate.net/publication/389309492_Draft_genome_of_the_endangered_visayan_spotted_deer_Rusa_alfredi_a_Philippine_endemic_species
Aksyon Radyo Iloilo interview with Philippine Genome Center Visayas: https://www.facebook.com/PGCVisayas/posts/pfbid029yugbJ6ANfSoSjH42Nrm9kDa5EwRFcGPo75dP7kpSxogv9GKV8kZpG1hZnouSxDQl
13 notes
·
View notes
Text
As much as I dislike how science works, I love all the public data available in databases! It's pretty cool being able to download and see on your own computer a 10000 yr old guy's genome sequence
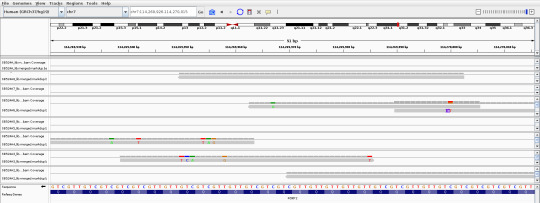
Cheddar man my beloved <3
This is a piece of FOXP2, a gene involved in speech and language development, very important for our ability to speak.
Article that sequenced his genome: https://www.nature.com/articles/s41559-019-0871-9 (unfortunately not public 😔)
ENA project ID: PRJEB31249 (https://www.ebi.ac.uk/ena/browser/view/PRJEB31249?show=reads) if you want to download it it's the SB524A to SB524A8 BAM & BAI files, it's a bit tricky to find on the paper.
#you don't even need a powerful computer to do it mine certainly isn't#just a few gb of space#genomes are heavy#genomics#science#biology
9 notes
·
View notes
Text
Hello! I used to have this domain as a studyblr but since finishing my masters, I saw little point in continuing that route so I decided to start from scratch. So consider this a little introduction back into this site that feels completely strange to me despite growing up in the Tumblr golden era.
My name is Lis, I'm a PhD student in Scientific Archaeology focusing on the effects of animal domestication in Central Europe. I'm currently struggling to find a passion for a subject that I genuinely love so I'm hoping that coming back to this little corner of the internet will reignite my love for history and learning.
#studyblr#archaeology#history#ancient history#culture#anthropology#european history#ancient culture#genomics#genetics#phd diaries
78 notes
·
View notes
Text
Hidden for 40,000 Years: Scientists Discover “Odd” New Butterfly Species

By Pensoft Publishers
5 May 2025
A new butterfly species, Satyrium curiosolus, has been discovered in Alberta.
Isolated for millennia, it shows unique traits and faces conservation risks due to low genetic diversity.
In Canada’s Rocky Mountains, a modest yet remarkable butterfly has gone largely unnoticed by science for years.
With a wingspan ranging from one to one and a half inches, and wings that are brown on top and greyish brown with black spots underneath, this butterfly was long believed to be part of the Half-moon Hairstreak species (Satyrium semiluna).
However, researchers have now confirmed that the isolated hairstreak butterflies living in Blakiston Fan, within Waterton Lakes National Park in Alberta, represent a distinct species: Satyrium curiosolus, or the Curiously Isolated Hairstreak.
A recent study by an international team, published in ZooKeys, revealed the butterfly’s unique evolutionary background.
The findings were striking: Satyrium curiosolus has likely been isolated from its closest relatives for as long as 40,000 years, gradually developing distinct genetic and ecological traits.
The science behind the discovery

“Our whole-genome sequencing of S. curiosolus revealed strikingly low genetic diversity and exceptionally high levels of historical inbreeding compared to the geographically nearest S. semiluna populations in British Columbia and Montana, more than 400 km distant,” explains co-first author Zac MacDonald, a La Kretz postdoctoral researcher at the University of California Los Angeles Institute of the Environment and Sustainability.
Although the population is small, genetic evidence indicates that S. curiosolus has remained a stable and independent lineage for tens of thousands of years.
“Like the Channel Island Fox, S. curiosolus may have purged some of its harmful recessive genetic variation through a long, gradual history of inbreeding, allowing it to persist as a small and completely isolated population today,” MacDonald adds.
Satyrium curiosolus is found in a distinct habitat unlike any other population of S. semiluna that we know of.
While its relatives thrive in sagebrush steppe, S. curiosolus occupies a single alluvial fan that is more accurately described as prairie-grassland, where it associates with different plants and ant species.
Satyrium curiosolus relies exclusively on silvery lupine (Lupinus argenteus) for larval development, a plant not known to be used by S. semiluna populations in British Columbia.
“Furthermore, we recently discovered that S. curiosolus larvae have mutualistic relationships with a particular species of ant (Lasius ponderosae), which has not been observed in other S. semiluna populations,” says James Glasier of the Wilder Institute/Calgary Zoo, who was also part of the study.
Satyrium curiosolus caterpillars provide the Lasius ants with a sugary excretion called honeydew to eat, while in return, the ants protect the caterpillar from parasites and predators.
Caterpillars also retreat into ant galleries when disturbed, or when it gets too hot out, and adult females have been observed laying eggs right near the entrances to Lasius colonies under Silvery Lupines.
Why it matters

The recognition of S. curiosolus as a species has important implications, highlighting its unique evolutionary trajectory and emphasizing an urgent need for tailored conservation strategies.
Satyrium curiosolus faces a somewhat unique challenge: its long-term isolation has resulted in very low genetic diversity, which means that the species has a reduced potential to adapt to changing climatic conditions.
While conservationists often consider genetic rescue — introducing individuals from related populations to boost genetic diversity — as a solution to low genetic diversity, the distinctiveness of S. curiosolus raises concerns about potential outbreeding depression when mixed with S. semiluna.
It is likely that the two species are not even reproductively compatible, meaning S. curiosolus may be on its own.
Conservation efforts must now consider new solutions, such as establishing additional S. curiosolus populations, to help this butterfly persist as climate change threatens ecological change at Blakiston Fan.
A case study in genomics and conservation

“The discovery of S. curiosolus is a powerful demonstration of how genomics is revolutionizing taxonomy and conservation,” remarked co-first author Julian Dupuis, an Assistant Professor in the Department of Entomology at University of Kentucky.
“While traditional taxonomic methods often rely on morphology alone, our study underscores the importance of integrating genomic and ecological data to uncover hidden diversity.
With the rise of genomic tools, previously unrecognized species like S. curiosolus are being discovered, highlighting the need for conservation strategies that account for cryptic biodiversity,” Dupuis adds.
Collaboration in conservation
Our studies on S. curiosolus and S. semiluna highlight the importance of collaboration between academic scientists, nonprofit organizations, and conservation managers.
All of this work was made possible through partnerships between academic researchers, Parks Canada, and the Wilder Institute/Calgary Zoo.
By combining expertise in genomics, field ecology, and conservation management, we were able to produce findings that not only reshape our understanding of biodiversity but also provide actionable insights for species protection.
Moving forward, these interdisciplinary collaborations will be critical for tackling complex conservation challenges and ensuring the long-term survival of species like S. curiosolus,” added MacDonald.
The future of Satyrium curiosolus

Recognizing S. curiosolus as a distinct species is just the beginning, the researchers say.
Future research should explore its evolution and interactions with other species, like host plants and ants.
Additionally, long-term monitoring by Parks Canada and the Wilder Institute/Calgary Zoo will be essential to assess how this species copes with climate change and what conservation actions are appropriate.
“This is a wonderful example of how such monitoring can connect diverse approaches and impactful answers to a simple question like ‘that’s odd – why is it there?’”, says anchor author Felix Sperling, a professor at the University of Alberta and curator of the U of A’s Strickland Museum of Entomology.
“For now, the Curiously Isolated Hairstreak reminds us that even the smallest and most overlooked species can hold extraordinary scientific and conservation significance,” the researchers say in conclusion.
#Satyrium curiosolus#butterfly#alberta#canada#rocky mountains#Satyrium semiluna#Curiously Isolated Hairstreak#ZooKeys#silvery lupine#genomics#field ecology#conservation management#conservation#Parks Canada#Wilder Institute/Calgary Zoo
6 notes
·
View notes
Text
Free online courses for bioinformatics beginners
🔬 Free Online Courses for Bioinformatics Beginners 🚀
Are you interested in bioinformatics but don’t know where to start? Whether you're from a biotechnology, biology, or computer science background, learning bioinformatics can open doors to exciting opportunities in genomics, drug discovery, and data science. And the best part? You can start for free!
Here’s a list of the best free online bioinformatics courses to kickstart your journey.
📌 1. Introduction to Bioinformatics – Coursera (University of Toronto)
📍 Platform: Coursera 🖥️ What You’ll Learn:
Basic biological data analysis
Algorithms used in genomics
Hands-on exercises with biological datasets
🎓 Why Take It? Ideal for beginners with a biology background looking to explore computational approaches.
📌 2. Bioinformatics for Beginners – Udemy (Free Course)
📍 Platform: Udemy 🖥️ What You’ll Learn:
Introduction to sequence analysis
Using BLAST for genomic comparisons
Basics of Python for bioinformatics
🎓 Why Take It? Short, beginner-friendly course with practical applications.
📌 3. EMBL-EBI Bioinformatics Training
📍 Platform: EMBL-EBI 🖥️ What You’ll Learn:
Genomic data handling
Transcriptomics and proteomics
Data visualization tools
🎓 Why Take It? High-quality training from one of the most reputable bioinformatics institutes in Europe.
📌 4. Introduction to Computational Biology – MIT OpenCourseWare
📍 Platform: MIT OCW 🖥️ What You’ll Learn:
Algorithms for DNA sequencing
Structural bioinformatics
Systems biology
🎓 Why Take It? A solid foundation for students interested in research-level computational biology.
📌 5. Bioinformatics Specialization – Coursera (UC San Diego)
📍 Platform: Coursera 🖥️ What You’ll Learn:
How bioinformatics algorithms work
Hands-on exercises in Python and Biopython
Real-world applications in genomics
🎓 Why Take It? A deep dive into computational tools, ideal for those wanting an in-depth understanding.
📌 6. Genomic Data Science – Harvard Online (edX) 🖥️ What You’ll Learn:
RNA sequencing and genome assembly
Data handling using R
Machine learning applications in genomics
🎓 Why Take It? Best for those interested in AI & big data applications in genomics.
📌 7. Bioinformatics Courses on BioPractify (100% Free)
📍 Platform: BioPractify 🖥️ What You’ll Learn:
Hands-on experience with real datasets
Python & R for bioinformatics
Molecular docking and drug discovery techniques
🎓 Why Take It? Learn from domain experts with real-world projects to enhance your skills.
🚀 Final Thoughts: Start Learning Today!
Bioinformatics is a game-changer in modern research and healthcare. Whether you're a biology student looking to upskill or a tech enthusiast diving into genomics, these free courses will give you a strong start.
📢 Which course are you excited to take? Let me know in the comments! 👇💬
#Bioinformatics#FreeCourses#Genomics#BiotechCareers#DataScience#ComputationalBiology#BioinformaticsTraining#MachineLearning#GenomeSequencing#BioinformaticsForBeginners#STEMEducation#OpenScience#LearningResources#PythonForBiologists#MolecularBiology
9 notes
·
View notes
Text
Scientists sequence DNA from 3,600-year-old cheese found in Bronze Age graves, revealing ancient kefir-making practices
- By Nuadox Crew -
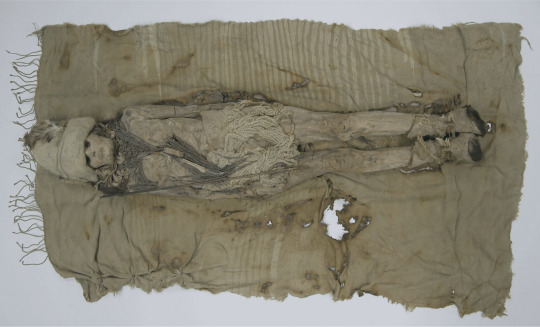
Scientists have sequenced DNA from 3,600-year-old cheese found in graves of the Xiaohe people, a Bronze Age culture in northwest China’s Taklamakan Desert.
The cheese, scattered on the heads and necks of mummified remains, offers insights into ancient cheesemaking and microbial practices. DNA analysis showed that the Xiaohe people made kefir cheese using goat and cattle milk without mixing different animal milks, a technique still used today.
The research reveals the presence of probiotic bacteria similar to modern kefir, showing how microbial strains evolved over time. This discovery challenges the belief that kefir originated solely in the Caucasus Mountains, pointing instead to Tibet as a key origin.
The study opens new possibilities for understanding ancient human culture, technology, and microbial use, with scientists noting that ancient microbial DNA studies were unimaginable a decade ago.

Image: Researchers extracted animal and microbial DNA from the kefir cheese found on the Tarim Basin mummies. Credit: Yimin Yang.
Header image: A mummy from the Xiaohe cemetery with dairy remains scattered around its neck. Credit: Li Wenying, Xinjiang Cultural Relics and Archaeology Institute (a grey filter was added).
Read more at CNN/Microsoft Start
Scientific paper: Bronze Age cheese reveals human-Lactobacillus interactions over evolutionary history, Cell (2024). DOI: 10.1016/j.cell.2024.08.008. www.cell.com/cell/fulltext/S0092-8674(24)00899-7
Read Also
Rights of the dead and the living clash when scientists extract DNA from human remains
9 notes
·
View notes
Text

Meet the unsung contributor to revolutionary breakthroughs in treating polio, cancer, HPV, and even COVID-19: Henrietta Lacks. Born in 1920 Roanoke, Virginia, Henrietta's mother Eliza died when she was only four, and she was ultimately raised by her maternal grandfather in Clover, Virginia. Henrietta worked as a tobacco farmer and attended a segregated school until the age of 14, when she gave birth to a son, Lawrence. A daughter, Elsie, was born three years later --to compound the family's difficulties, Elsie had cerebral palsy and epilepsy. Henrietta and her now-husband David Lacks moved to Turner Station (now Dundalk), Maryland where David had landed a job with a nearby steel plant. At the time Turner Station was one of the oldest African-American communities in Baltimore County and there was sufficient community support for the family to buy a house and produce three more children.
In 1951 at the age of 31, Henrietta died at Johns Hopkins Hospital of cervical cancer, mere months after the birth of the family's youngest son. But before her death --and without her or her family's consent-- during a biopsy two tumour cell samples were taken from Henrietta's cervix and sent to Johns Hopkins researchers. Hernietta's cells carried a unique trait: an ability to rapidly multiply, producing a new generation every 24 hours; a breakthrough that no other human cell had achieved. Prior to this discovery, only cells that had been transformed by viruses or genetic mutations carried such a characteristic. With the prospect of now being able to work with what amounted to the first-ever naturally-occurring immortal human cells, researchers created a patent on the HeLa cell line but hid the donor's true identity under a fake name: Helen Lane.
It is no exaggeration to state that in the 70 years since her death, Henrietta's cells have been bought, sold, packaged, and shipped by thousands of laboratories; with her cells being used as a baseline in as many as 74,000 different studies (including some Nobel Prize winners). Her cells have even been sent into space to study the effects of microgravity, and were instrumental in the Human Genome Project. While no actual law (or even a code of ethics) necessarily required doctors to ask permission before taking tissue from a terminal patient, there was a very clear Maryland state law on the books that forbade tissue removal from the dead without permission, throwing the situation into something of a legal grey area. However because Henrietta was poor, minimally educated, and Black, this standard was quietly (and easily) circumvented and she was never recognized for her monumental contributions to science and medicine ...and her family was never compensated. The family remained unaware of Henrietta's contribution until 1975, when the HeLa line's provenance finally became public. Henrietta had been buried in an unmarked grave in the family cemetery in Clover, Virginia but in 2010 a new headstone was donated and dedicated, acknowledging her phenomenal contribution. That same year the John Hopkins Institute for Clinical and Translational Research established a new Henrietta Lacks Memorial lecture series. A statue of Lacks was commissioned in 2022, to be erected in Lacks's birthplace of Roanoke, Virginia --pointedly replacing a previous statue of Confederate Gen. Robert E. Lee, which had been removed following nationwide protests over the murder of George Floyd.
Dive into The Immortal Life of Henrietta Lacks by Rebecca Skloot, originally published in 2011 and subsequently adapted into an HBO movie in 2017, starring Oprah Winfrey as Henrietta's daughter Deborah and Renee Elise Goldberry as Henrietta. (And yes, this book has been challenged and banned in more than one school district.)
#black lives matter#henrietta lacks#johns hopkins#cell biology#hela#stem cell#translational research#genomics#teachtruth#dothework
65 notes
·
View notes
Text




Trying to find a balance
#studyblr#study motivation#grad school life#studying#reading#genomics#anthropology#academia#chaotic academia#chaotic academic aesthetic#academic#Spotify
16 notes
·
View notes