#actinomycetota
Text
Actinomycetota vs. Thermomicrobiota
Actinomycetota propaganda here
Thermomicrobiota propaganda here

31 notes
·
View notes
Text
Applied Microbiology, Vol. 4, Pages 1268-1282: Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil
Composting is an environmentally friendly process, turning animal waste into fertilizer. Chicken litter compost (CLC) improves soil properties, increasing crop yields. However, the CLC effect on the soil microbiome is understudied. This study aimed to compare bacteriobiome diversity in fallow arable Chernozem with and without CLC addition in a field experiment in the Novosibirsk region, Russia, using 16S #rRNA gene metabarcoding. Pseudomonadota, Actinomycetota and Acidobacteriota were the most OTU-rich phyla, together accounting for >50% of the total number of sequence reads. CLC-related shifts in the bacteriobiome structure occurred at all taxonomic levels: the Bacillota abundance was 10-fold increased due to increased Bacilli, both being indicator taxa for the CLC-soil. The main Actinomycetota classes were the indicators for the CLC-soil (Actinobacteria) and no-CLC soil (Thermoleophilia, represented Gaiella). Both Bacillota and Actinomycetota phyla were the ultimate constituents of the CLC added, persisting in the soil for five months of fallowing. The no-CLC soil indicator phyla were Acidobacteriota (represented by Acidobacteria_Group3) and Verrucomicrobiota. Future metabarcoding studies of chicken litter application in agricultural soils, including cropped studies, should address the soil microbiome at the species/strain levels in more detail, as well as how it is affected by specific crops, preferably accompanied by a direct methodology revealing the microbiota functions. https://www.mdpi.com/2673-8007/4/3/87?utm_source=dlvr.it&utm_medium=tumblr
0 notes
Text
Antifungal mechanism of volatile compounds emitted by Actinomycetota Paenarthrobacter ureafaciens from a disease-suppressive soil on Saccharomyces cerevisiae
Pubmed: http://dlvr.it/SwfH2N
0 notes
Text
Prodigiosin, Anti-Cancer Substance

image from the Internet
Prodigiosin is a natural red pigment with a methoxy-pyrrole skeleton structure of 3 pyrrole type rings. Prodigiosin is a secondary metabolite of Serratia marcescens, Actinomycetota, and so on. It is responsible for the pink tint occasionally found in grime that accumulates on porcelain surfaces such as bathtubs, sinks, and toilet bowls. As a natural pigment derived from microorganisms, prodigiosin has broad application prospects and market value in the fields of medicine, environmental management, textile dyes and even cosmetics.

image source: KingDraw
Facebook: KingDraw_chem
FB group: KingDraw Q&A
Twitter: KingDraw_chem
YouTube: KingDraw
Medium: KingDraw
Quora: KingDraw
You can also email [email protected] for more software tutorials :)
#chemisry#organicchmistry#compound#science#molecule#online tutoring#research chemicals#anticancerfood
0 notes
Text
Actinomycetota
Group: Terrabacteria
Gram-stain: Positive
Etymology: For Actinomyces bovis. From the Greek "aktis", meaning "ray", and "myketes", meaning "fungus". Some bacteria in this phylum form fungus-like branched colonies, hence the name: "ray fungus". Note the similarity to mycelium networks.
About: Actinomycetota contains tuberculosis (Mycobacterium tuberculosis), the leading cause of death by bacterial infection. It is also the second leading cause of death by any infectious disease, only surpassed by COVID-19. Many other members of the genus Mycobacterium are also pathogenic, such as Mycobacterium leprae, which is responsible for Hansen's disease (better known as leprosy).
But Actinomycetota is far from all bad. In fact, it is possibly the most studied and used bacterial phylum in terms of medical, agricultural, environmental, and biotechnological applications. Actinomycetota play a huge role in nutrient cycling in soil ecosystems, making them crucial for maintaining the health of soil. They are therefore a pillar of modern agriculture. Some Actinomycetota are nitrogen-fixing, and can be symbiotic with trees. The phylum is also a source of antibiotics, especially with the genus Streptomyces: these bacteria naturally produce several antimicrobials (including streptomycin), and thus have use both in medicine and as a natural pesticide against other microorganisms in the soil.
Actinomycetota colonies have long been noted to resemble those of fungi, and early researchers originally believed that this phylum was a fungi. Indeed, Actinomycetota and fungi share similar roles, being largely decomposers. However, the much smaller Actinomycetota fill their own ecological niche.
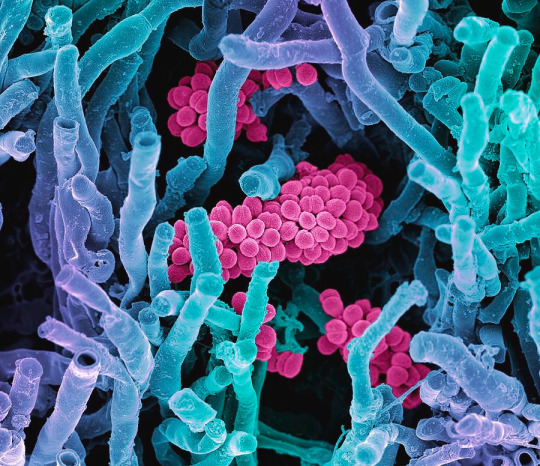
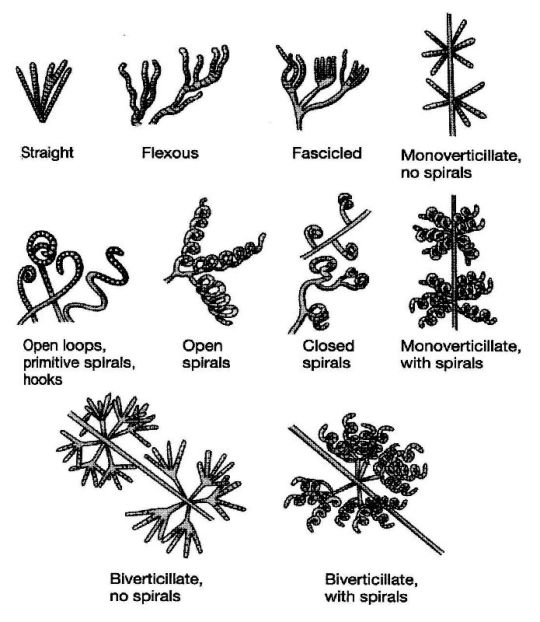
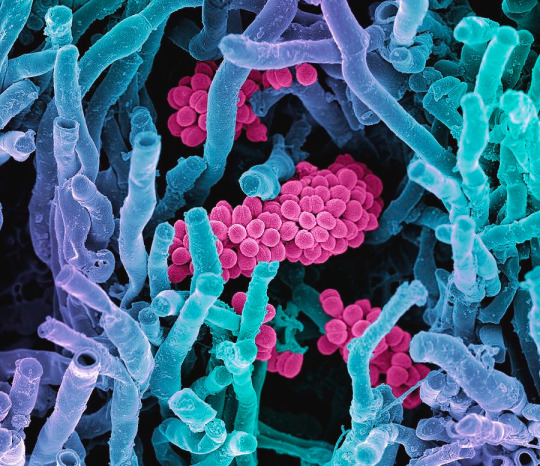
In terms of reproduction, Actinomycetota and fungi are especially similar. Actinomycetota produce spores, but not just endospores: they also form spores using external structures called "hyphae". Hyphae are branched, filamentous structures that make up what is called a "mycelium", and can have a variety of shapes. Many hyphae, including those in Actinomycetota, are reproductive structures that subdivide to create spores. This is a process ubiquitous among fungi but limited to pretty much only the Actinomycetota phylum of bacteria. The below image depicts some possible hyphae structures in Streptomyces bacteria:
In addition to all this, Actinomycetota may contain the oldest living organisms on earth, which could have been frozen in Siberian or Antarctic ice for thousands (or millions) of years. While Actinomycetota do produce durable endospores, it is also theorized that they may also be able to survive in a frozen but non-dormant state: not reproducing, but still metabolizing, and thus being able to repair their DNA.
While the theory is speculative, evidence of metabolic activity has been detected from soil samples after more than 600,000 years of permafrost, studied under ambient conditions (i.e. not thawed to trigger endospore germination). We have actually cultivated Actinomycetota from other ancient permafrost samples, such as 28,000 year old mammoth poop.
6 notes
·
View notes
Text
Phylum Bracket Announcement!
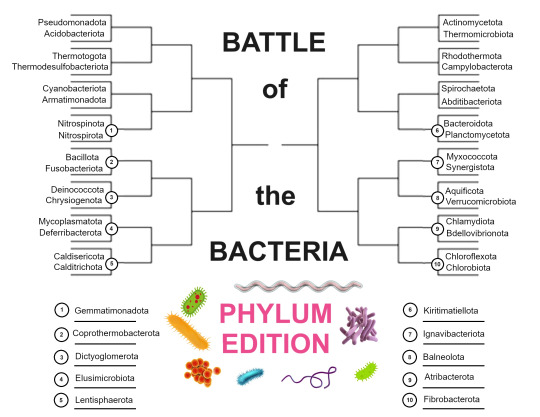
42 bacterial phyla, from two sources: Oren & Garrity (2021), and the list of recognized bacterial phyla under the ICNP as of February 2024. The Oren & Garrity list is more expansive, as it includes bacterial phyla that have been well studied, but not cultivated in the lab, while the ICNP restricts itself to bacteria that have been successfully cultured. 36 phyla were on both lists, 4 were exclusive to Oren & Garrity, and 2 were exclusive to the ICNP. Two phyla from the Oren & Garrity list were excluded for not being bacteria, as the list was of prokaryotic phyla generally.
The first round will be between 32 of the phyla, while ten sit out. Then the second round, which will be called round 1.5, will pair ten of the winners from the first round against those ten. Thus, after round 1.5, there will be 16 bacterial phyla remaining, and the tournament proceeds as is standard from there.
Here is a list of the phyla, organized (roughly) by clade and superphylum. Clades/superphyla are in bold and phylum names are in italics for clarity.
Terrabacteria
Abditibacteriota
Actinomycetota
Armatimonadota
Bacillota
Chloroflexota
Cyanobacteria
Mycoplasmatota
Thermomicrobiota
Hydrobacteria
Elusimicrobiota
Spirochaetota
Proteobacteria
Acidobacteriota
Aquificota
Bdellovibrionota
Campylobacterota
Chrysiogenota
Deferribacterota
Myxococcota
Nitrospinota
Nitrospirota
Pseudomonadota
Thermodesulfobacteriota
PVC Group
Chlamydiota
Lentisphaerota
Kirimatiellota
Planctomycetota
Verrucomicrobiota
FCB Group
Bacteroidota
Balneolota
Chlorobiota
Fibrobacterota
Gemmatimonadota
Ignavibacteriota
Rhodothermota
Thermotogida
Synergistetes
Atribacterota
Synergistota
Thermocalda
Caldisericota
Calditrichota
Coprothermobacterota
Dictyoglomerota
Thermotogota
Other
Deinococcota
Fusobacteriota
Propaganda is still underway, but look out for an announcement of the first battles soon! I am planning to do round 1 in two parts over two weeks, with 8 battles in each week. Get ready: the girls are fighting !!!
66 notes
·
View notes
Text
Round 1 is Complete!

In the second part of the first round, Actinomycetota, Rhodothermota, Spirochaetota, Planctomycetota, Myxococcota, Verrucomicrobiota, Bdellovibrionota and Chloroflexota were victorious! We will be saying goodbye to Thermomicrobiota, Campylobacterota, Abditibacterota, Bacteroidota, Synergistota, Chlamydiota, and Chlorobiota.
The list of bacteria that will proceed to Round 2 will be determined after Round 1.5, which will consist of the following 10 battles!
Nitrospirota vs. Gemmatimonadota
Bacillota vs. Coprothermobacterota
Deinococcota vs. Dictyoglomerota
Mycoplasmatota va. Elusimicrobiota
Caldisericota vs. Lentisphaerota
Planctomycetota vs. Kiritimatiellota
Myxococcota vs. Ignavibacteriota
Verrucomicrobiota vs. Balneolota
Bdellovibrionota vs. Atribacterota
Chloroflexota vs. Fibrobacterota
I have a lot of propaganda left to write for the new bacteria in this round, so I expect it to be about a week before that round is ready.
THANK YOU all for voting! The most popular poll this round got over 200 votes, and I am so happy to see that kind of engagement over microbiology. I myself have learned SO MUCH about bacteria through this project and hope to learn more about the field afterwards... honestly this whole tumblr taxonomy community has been really inspiring!
THANK YOU as well to: @0ctogus! They proof-read basically ALL of the propaganda that hits your dash (with the exception of some early examples), and were generally super helpful as a source of quality-control and peer-review, from someone who has a much better knowledge of biology than I. It didn't feel right to finish this round off without mentioning their contributions!
And that concludes Round 1! See you all soon with more Bacteria Facts.
16 notes
·
View notes
Text
Chloroflexota
Group: Terrabacteria
Gram-stain: Varied
Etymology: For Chloroflexus aurantiacus. From the Greek "chloros", meaning "yellowish green", and Latin "flexus", meaning "bending", for their green color.
About: Chloroflexota, known for containing the "green non-sulfur bacteria", is a highly diverse and ubiquitous phylum. They exhibit a variety of oxygen tolerances, and may be aerobic, anaerobic, or somewhere in between. Members of Chloroflexota can be thermophiles or mesophiles, living in a range of environments such as hot springs, sea-floor sediments, soil, and anaerobic sludge bioreactors. They are largely chemoheteroorganotrophic, with several members also capable of photoautotrophy. Despite their prevalence, Chloroflexota have limited cultivability, and are therefore still quite understudied. The species Thermoflexus hugenholtzii are especially picky, with the narrowest growth-temperature range (in culture) of any known prokaryote (67.5°- 75° C).
On the Gram stain, Chloroflexota show varied results. Most are monoderms, having only one cell membrane, but many still stain gram-negative. This is due to the unique composition of their cell walls (one factor of which is the higher presence of a molecule called "pseudopeptidoglycan", rather than being primarily peptidoglycan). There are also plenty of gram-positive, spore-producing Chloroflexota. These share similarities with Actinomycetota and fungi, since they produce spores using hyphae, and form mycelium.

The name "green non-sulfur bacteria" is associated with the family Chloroflexaceae, in the order Chloroflexales. The Chloroflexales are known as the "filamentous anoxygenic phototrophic bacteria", or FAPs, for their style of photosynthesis that does not produce oxygen (in contrast to Cyanobacteriota and plants). There are "red FAPs" and "green FAPs", with the green FAPs constituting the green non-sulfur bacteria, in the family Chloroflexaceae.
Green non-sulfur bacteria share many similarities with their counterparts, the green sulfur bacteria (Chlorobiota), despite being distantly related. Both groups form the same antennae structures, filled with bacteriochlorophyll-containing chlorosomes that color them green. Chloroflexaceae, however, are not primarily photosynthetic. Instead, they are facultative anaerobes who tend to use a chemoheterotrophic metabolism in the presence of oxygen, and a photoautotrophic metabolism in its absence.
Another interesting family of Chloroflexota are the Dehalococcoidaceae, because they are involved in halogen-cycling. The bacteria are organohalide-respiring (halogens are reactive elements belonging to the group containing fluorine and chlorine, and an organohalide is an organic compound with a carbon-halogen bond). Thanks to this style of respiration, Dehalococcoidaceae are able to thrive in chlorinated environments. This makes them useful in the bioremediation of chlorine-contaminated ecosystems. Also, they can produce metabolites that smell like garlic.
10 notes
·
View notes
Text
Microorganisms, Vol. 12, Pages 397: Analysis of Culturable Bacterial Diversity of Pangong Tso Lake via a 16S #rRNA Tag Sequencing Approach
The Pangong Tso lake is a high-altitude freshwater habitat wherein the resident microbes experience unique selective pressures, i.e., high radiation, low nutrient content, desiccation, and temperature extremes. Our study attempts to analyze the diversity of culturable bacteria by applying a high-throughput amplicon sequencing approach based on long read technology to determine the spectrum of bacterial diversity supported by axenic media. The phyla Pseudomonadota, Bacteriodetes, and Actinomycetota were retrieved as the predominant taxa in both water and sediment samples. The genera Hydrogenophaga and Rheinheimera, Pseudomonas, Loktanella, Marinomonas, and Flavobacterium were abundantly present in the sediment and water samples, respectively. Low nutrient conditions supported the growth of taxa within the phyla Bacteriodetes, Actinomycetota, and Cyanobacteria and were biased towards the selection of Pseudomonas, Hydrogenophaga, Bacillus, and Enterococcus spp. Our study recommends that media formulations can be finalized after analyzing culturable diversity through a high-throughput sequencing effort to retrieve maximum species diversity targeting novel/relevant taxa. https://www.mdpi.com/2076-2607/12/2/397?utm_source=dlvr.it&utm_medium=tumblr
0 notes