#Animal Genetics and Genomics
Explore tagged Tumblr posts
Photo

The Solanum americanum genome has been used to discover immune receptors that detect potato late blight pathogen effectors. The discovery could lead to the development of new, more effective ways to fight the late blight pathogen.
#Genomics#Plant genetics#Biomedicine#general#Human Genetics#Cancer Research#Agriculture#Gene Function#Animal Genetics and Genomics#fault#Solanum americanum#genome#immune receptors#late blight pathogen#effectors#development
1 note
·
View note
Text

Largest animal genome sequenced — and just 1 chromosome is the size of the entire human genome
Scientists sequenced the largest known animal genome in a species of lungfish — ancient fish that breathe air.
Scientists have sequenced the largest known animal genome — and it's 30 times bigger than the human genome. The genome belongs to the South American lungfish (Lepidosiren paradoxa), a primeval, air-breathing fish that "hops" onto land from the water using weird, limb-like fins. The fish's DNA code expanded dramatically over the past 100 million years of evolutionary history, racking up the equivalent of one human genome every 10 million years, researchers found. The findings could shed light on how genomes expand across the tree of life...
Read more: Largest animal genome sequenced — and just 1 chromosome is the size of the entire human genome | Live Science
971 notes
·
View notes
Text

A genome-based phylogeny for Mollusca is concordant with fossils and morphology
Abstract
Extreme morphological disparity within Mollusca has long confounded efforts to reconstruct a stable backbone phylogeny for the phylum. Familiar molluscan groups—gastropods, bivalves, and cephalopods—each represent a diverse radiation with myriad morphological, ecological, and behavioral adaptations. The phylum further encompasses many more unfamiliar experiments in animal body-plan evolution. In this work, we reconstructed the phylogeny for living Mollusca on the basis of metazoan BUSCO (Benchmarking Universal Single-Copy Orthologs) genes extracted from 77 (13 new) genomes, including multiple members of all eight classes with two high-quality genome assemblies for monoplacophorans. Our analyses confirm a phylogeny proposed from morphology and show widespread genomic variation. The flexibility of the molluscan genome likely explains both historic challenges with their genomes and their evolutionary success.
Read the paper here:
A genome-based phylogeny for Mollusca is concordant with fossils and morphology | Science
132 notes
·
View notes
Text
The longest genome of all the animals on Earth belongs not to a giant, or a cognitively advanced critter, but a writhing, water-dwelling creature seemingly frozen in time, right at the cusp of evolving into a beast that can live on land. These are the lungfish, a class of freshwater vertebrates whose peculiar characteristics are reflected in a colossal genetic code. Able to breathe both air and water, with limb-like fins, and a well-developed skeletal architecture, these strange ancient creatures are thought to is thought to share a common ancestor with all four-limbed vertebrates known as tetrapods.
Continue Reading.
302 notes
·
View notes
Text
A protein found in human sweat may protect against Lyme disease
New Post has been published on https://thedigitalinsider.com/a-protein-found-in-human-sweat-may-protect-against-lyme-disease/
A protein found in human sweat may protect against Lyme disease


Lyme disease, a bacterial infection transmitted by ticks, affects nearly half a million people in the United States every year. In most cases, antibiotics effectively clear the infection, but for some patients, symptoms linger for months or years.
Researchers at MIT and the University of Helsinki have now discovered that human sweat contains a protein that can protect against Lyme disease. They also found that about one-third of the population carries a genetic variant of this protein that is associated with Lyme disease in genome-wide association studies.
It’s unknown exactly how the protein inhibits the growth of the bacteria that cause Lyme disease, but the researchers hope to harness the protein’s protective abilities to create skin creams that could help prevent the disease, or to treat infections that don’t respond to antibiotics.
“This protein may provide some protection from Lyme disease, and we think there are real implications here for a preventative and possibly a therapeutic based on this protein,” says Michal Caspi Tal, a principal research scientist in MIT’s Department of Biological Engineering and one of the senior authors of the new study.
Hanna Ollila, a senior researcher at the Institute for Molecular Medicine at the University of Helsinki and a researcher at the Broad Institute of MIT and Harvard, is also a senior author of the paper, which appears today in Nature Communications. The paper’s lead author is Satu Strausz, a postdoc at the Institute for Molecular Medicine at the University of Helsinki.
A surprising link
Lyme disease is most often caused by a bacterium called Borrelia burgdorferi. In the United States, this bacterium is spread by ticks that are carried by mice, deer, and other animals. Symptoms include fever, headache, fatigue, and a distinctive bulls-eye rash.
Most patients receive doxycycline, an antibiotic that usually clears up the infection. In some patients, however, symptoms such as fatigue, memory problems, sleep disruption, and body aches can persist for months or years.
Tal and Ollila, who were postdocs together at Stanford University, began this study a few years ago in hopes of finding genetic markers of susceptibility to Lyme disease. To that end, they decided to run a genome-wide association study (GWAS) on a Finnish dataset that contains genome sequences for 410,000 people, along with detailed information on their medical histories.
This dataset includes about 7,000 people who had been diagnosed with Lyme disease, allowing the researchers to look for genetic variants that were more frequently found in people who had had Lyme disease, compared with those who hadn’t.
This analysis revealed three hits, including two found in immune molecules that had been previously linked with Lyme disease. However, their third hit was a complete surprise — a secretoglobin called SCGB1D2.
Secretoglobins are a family of proteins found in tissues that line the lungs and other organs, where they play a role in immune responses to infection. The researchers discovered that this particular secretoglobin is produced primarily by cells in the sweat glands.
To find out how this protein might influence Lyme disease, the researchers created normal and mutated versions of SCGB1D2 and exposed them to Borrelia burgdorferi grown in the lab. They found that the normal version of the protein significantly inhibited the growth of Borrelia burgdorferi. However, when they exposed bacteria to the mutated version, twice as much protein was required to suppress bacterial growth.
The researchers then exposed bacteria to either the normal or mutated variant of SCGB1D2 and injected them into mice. Mice injected with the bacteria exposed to the mutant protein became infected with Lyme disease, but mice injected with bacteria exposed to the normal version of SCGB1D2 did not.
“In the paper we show they stayed healthy until day 10, but we followed the mice for over a month, and they never got infected. This wasn’t a delay, this was a full stop. That was really exciting,” Tal says.
Preventing infection
After the MIT and University of Helsinki researchers posted their initial findings on a preprint server, researchers in Estonia replicated the results of the genome-wide association study, using data from the Estonian Biobank. These data, from about 210,000 people, including 18,000 with Lyme disease, were later added to the final Nature Communications study.
The researchers aren’t sure yet how SCGB1D2 inhibits bacterial growth, or why the variant is less effective. However, they did find that the variant causes a shift from the amino acid proline to leucine, which may interfere with the formation of a helix found in the normal version.
They now plan to investigate whether applying the protein to the skin of mice, which do not naturally produce SCGB1D2, could prevent them from being infected by Borrelia burgdorferi. They also plan to explore the protein’s potential as a treatment for infections that don’t respond to antibiotics.
“We have fantastic antibiotics that work for 90 percent of people, but in the 40 years we’ve known about Lyme disease, we have not budged that,” Tal says. “Ten percent of people don’t recover after having antibiotics, and there’s no treatment for them.”
“This finding opens the door to a completely new approach to preventing Lyme disease in the first place, and it will be interesting to see if it could be useful for preventing other types of skin infections too,” says Kara Spiller, a professor of biomedical innovation in the School of Biomedical Engineering at Drexel University, who was not involved in the study.
The researchers note that people who have the protective version of SCGB1D2 can still develop Lyme disease, and they should not assume that they won’t. One factor that may play a role is whether the person happens to be sweating when they’re bitten by a tick carrying Borrelia burgdorferi.
SCGB1D2 is just one of 11 secretoglobin proteins produced by the human body, and Tal also plans to study what some of those other secretoglobins may be doing in the body, especially in the lungs, where many of them are found.
“The thing I’m most excited about is this idea that secretoglobins might be a class of antimicrobial proteins that we haven’t thought about. As immunologists, we talk nonstop about immunoglobulins, but I had never heard of a secretoglobin before this popped up in our GWAS study. This is why it’s so fun for me now. I want to know what they all do,” she says.
The research was funded, in part, by Emily and Malcolm Fairbairn, the Instrumentarium Science Foundation, the Academy of Finland, the Finnish Medical Foundation, the Younger Family, and the Bay Area Lyme Foundation.
#000#Analysis#Animals#antibiotic#Antibiotics#antimicrobial#approach#Bacteria#Biological engineering#Biology#Broad Institute#Cells#communications#data#Delay#Disease#disruption#engineering#eye#factor#fatigue#Finland#Foundation#Full#genetic#Genetics#genome#growth#how#human
2 notes
·
View notes
Text
Lizards that once dwelled in forests but now slink around urban areas have genetically morphed to survive life in the city.
The species crested anole lizard has scales that have adapted to better cling to smooth surfaces like walls and windows and grew larger limbs to sprint across open areas like hot parking lots.
The study analyzed 96 Anolis cristatellus lizards, comparing the genetic makeup of forest-dwellers to those living in Puerto Rico's capital, San Juan, as well as the northern city of Arecibo and western city of Mayaguez. Scientists found that 33 genes within the lizard genome were repeatedly associated with urbanization.
#biology#evolution#lizard#puerto rico#animal adaptation#urban wildlife#deforestation#genetics#genome
6 notes
·
View notes
Text
My favorite detail about Jurassic Park is that it has a baked-in justification for any and all retcons it might need to make due to paleontology advancing forwards.
Because there is not a single dinosaur that has ever appeared in Jurassic Park.
Not one. Not in the books. Not in the movies. Not ever.
"Now what John Hammond and InGen did at Jurassic Park was to create genetically engineered theme park monsters." ~Alan Grant
Grant says that in a moment of cynicism. It's part of his arc for the film. But it's not inaccurate. What Jurassic Park has, what it's always had since the very first novel, are "Mostly Dinosaurs".
"And since the DNA is so old, it's full of holes! Now, that's where our geneticists take over!" ~Mr. DNA
It's impossible to recover a fully intact gene sequence from an ancient amber mosquito. Cloning a pure dinosaur would have been completely impossible, and so the park filled in the gene sequence with whatever works. Frog. Lizard. Bird. Whatever they need to get the result they are trying to get.
Every single dinosaur is a chimeric beast made up of mostly dinosaur and a bunch of other stuff that some scientists thought would achieve the appropriate dinosaur-like result.
"Nothing in Jurassic World is natural! We have always filled gaps in the genome with the DNA of other animals. And if the genetic code was pure, many of them would look quite different." ~Dr. Henry Wu
Which, from a writing perspective, is fucking genius. Because now you have a preset excuse for each and every plot hole your movie has.
Like. Why don't the raptors have feathers? Because of the chimera DNA.
Why do dilophosaurs spit venom? Because of the chimera DNA.
Why do T-Rexes have movement based vision? Oh, they don't. But Rexy does. Because of her chimera DNA.
Why is the Spinosaurus so fucking big? Because of the chimera DNA.
Why are the velociraptors mislabeled? Because Hammond's a dipshit.
Like. I've always marveled at the way Jurassic Park started out by giving itself a blanket excuse to be wrong about every single thing it ever said about the central attraction of its franchise. It's honestly beautiful, and allows the series a degree of immortality well into the era where we know better about its animals.
26K notes
·
View notes
Text
RAAAAH I HATE SHITTY SCI COMM This post is great & elaborates more on the actual project!!!
Support Red wolf conservation with organizations such as the Red Wolf Coalition, Wolf Conservation Center, and the National Wildlife Federation :)
https://www.nwf.org/Our-Work/Wildlife-Conservation/Red-Wolf
i've been trying to stay off of internet and i've been active on tumblr because I'm too exhausted to do things I normally enjoy. Anyway
Animal enjoyers are mad about the slightly edited wolves that Colossal Biosciences is claiming are "dire wolves." Lots of them didn't read the articles, which would provide more information. However, the journalism about this has been god-awful anyway.
The company is concurrently working on cloning endangered red wolves and figuring out how to bring red wolf/coyote hybrids back into the red wolf gene pool, as per the Time article about it. The project includes one of the biggest names in canid genomics and evolution including pertaining to red wolves, so I am optimistic that red wolves are probably the real aim of the project and the dire wolf bullshit is just a snazzy jurassic park style tagline to snare investors.
However the grift has grifted too close to the sun as according to washington post, trump is using "de-extinction" technology as an excuse to gut the endangered species act (i can't actually read the article unfortunately). The cost of this lie could be very high if the general public thinks that bringing back an extinct species can be easily done by just going into the DNA of an animal that looks sort of similar and tweaking it.
Also somehow, even more infuriating to me, this is going to eternally fuck up the perception of what a dire wolf actually was. As per wikipedia, Aencyon dirus was not closely related to any modern wolves. It is over 5 million years separate from them. It was essentially not a "wolf" at all. You might as well try to create a dire wolf by modifying a jackal or an African wild dog. You might as well call the dire wolf a dire jackal or a dire dog.
Dire wolves were not that much bigger than wolves. They were maybe 20% bigger and their size range overlaps with the northern-most wolves of today.
Even the articles critical of the supposed "de-extinction" are fucking it up! The not-actually-legit "dire wolf" puppies have white fur, and the journalists are uncritically repeating the idea that dire wolves were white, when that isn't something we know about them. The white fur is based off of the fantasy creature of the same name in Game of Thrones.
That's just flat-out embarrassing.
#animals#scicomm#science#genomics#genetics#conservation#the colossal bullshit#dire wolf#red wolf#save the red wolf
2K notes
·
View notes
Text
We do not know how virulent it was, but we do know that it still contained genes that were shared by an ancestral, animal-infecting relative of all human smallpox; thus, ancient smallpox would still have infected a wider range of animal hosts. The medieval smallpox genomes also allow the molecular clock to be refined, and it can be estimated that these strains shared a common ancestor with modern smallpox around 1,700 years ago. Smallpox, as a human disease in some form, is at least that old.⁷³
73. Both Aelius Aristides and Herodian observed animal deaths. Although such reports can be taken with a grain of salt, the possibility that ancient smallpox infected a wider range of boats is an important response to previous objections that the Antonine Plague could not have been smallpox on these grounds.
"Plagues Upon the Earth: Disease and the Course of Human History" - Kyle Harper
#book quotes#plagues upon the earth#kyle harper#nonfiction#virulent#genes#genetics#genome#infection#smallpox#medieval#aelius aristides#herodian#animal death#observation#salt#objection#antonine plague
1 note
·
View note
Text
Animal Genetics Market Projected to Reach $9.12 Billion by 2031
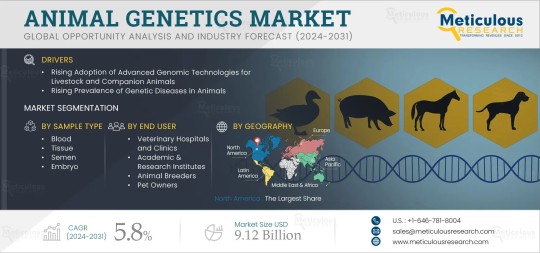
Meticulous Research®—a leading global market research company, published a research report titled ‘Animal Genetics Market Size, Share, Forecast, & Trends Analysis by Animal (Companion [Dogs, Cats] Livestock [Poultry, Porcine, Bovine]) Services (DNA Profile, Genetic Diseases, Traits) Sample Type (Blood, Tissue, Embryo), End User - Global Forecast to 2031.’
According to a recent publication from Meticulous Research®, the animal genetics market is projected to reach $9.12 billion by 2031, with a CAGR of 5.8% during the forecast period. Historically, crossbreeding—mating animals from different breeds to combine desirable traits—relied on physical characteristics without genetic insights. However, recent advancements have seen a surge in genetic testing, providing breeders with data on genetic profiles and trait-associated genes. This enables improved breeding and crossbreeding for better meat and milk production, and increased disease resistance.
Market growth is driven by the demand for precision breeding techniques, rising awareness about the benefits of genetic testing for disease detection, the adoption of advanced genomic technologies, government support for genetic research, and the prevalence of genetic diseases in animals. Emerging markets in Asia-Pacific and Latin America, along with the development of portable and affordable genetic testing solutions, present growth opportunities.
Key players in the animal genetics market include Animal Genetics, Inc., Zoetis Inc., Mars Petcare, Embark Veterinary, Inc., Neogen Corporation, Basepaws Inc., LABOKLIN GMBH & CO.KG, Generatio GmbH, Vetgen LLC, FarmLab Diagnostics, and EasyDNA.
Future Outlook
The animal genetics market is segmented based on testing services, sample type, animal type, end user, and geography:
Testing Services: Includes DNA profile testing, genetic traits testing, genetic disease testing, and other testing services. The genetic disease testing segment is expected to have the highest CAGR of 6.7% from 2024 to 2031 due to the need for early detection and management of infectious and metabolic diseases.
Sample Type: Segments include blood, tissue, semen, embryo, and others. The blood segment is projected to grow the fastest due to its convenience and reliability for genetic testing.
Animal Type: Divided into companion animals, livestock, and others. The companion animals segment is anticipated to hold the largest market share in 2024, driven by the global pet population and demand for pet health and breeding.
End User: Comprises veterinary hospitals and clinics, academic and research institutes, animal breeders, and pet owners. The animal breeders segment is expected to dominate in 2024 due to the demand for high-quality livestock and purebred companion animals.
Geographic Review
The report covers major regions including North America, Europe, Asia-Pacific, Latin America, and the Middle East & Africa. North America is expected to hold the largest market share in 2024, while emerging markets like India and China are projected to grow the fastest, driven by large exports, high production of animal-derived products, and increasing pet adoption.
Download Sample Report Here @ https://www.meticulousresearch.com/download-sample-report/cp_id=5863
Key Questions Answered
Which segments are experiencing high growth in the market?
What was the historical market size globally?
What are the market forecasts and estimates for 2024–2031?
What are the major drivers, restraints, opportunities, and challenges?
Who are the major players and what is the competitive landscape?
What are the recent developments and strategies of key players?
What are the geographical trends and high-growth regions?
Contact Us: Meticulous Research® Email- [email protected] Contact Sales- +1-646-781-8004 Connect with us on LinkedIn- https://www.linkedin.com/company/meticulous-research
#Animal Genetics Market#Animal Genetics#Pet DNA Testing#Animal Genomics#Molecular Breeding#Pet Genetics#Animal Breeding#Pet Breeding#Animal Genetic Engineering
0 notes
Text
Dog DNA Determines Guilt
It is surprising how inexpensive and accessible DNA testing has become. Many readers of Mere Inkling have submitted samples ourselves. But how many of us have had our pets tested? The science of genetics is quite recent. Gregor Mendel (1822-1884) is considered the father of such studies. As Britannica describes the history, “The word genetics was introduced in 1905 by English biologist William…

View On WordPress
#Animal Cruelty#Animals#C.S. Lewis#Dead Poets#DNA#Dogs#France#Genetics#Genome#Passports#Pets#Poetry#San Francisco#Toilets#Veterinarians
0 notes
Text
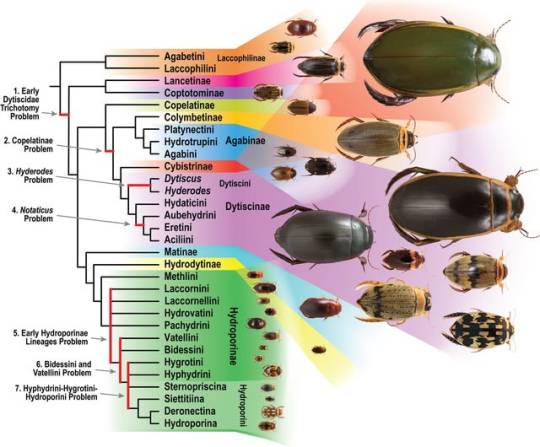
Whole genome shotgun phylogenomics resolve the diving beetle tree of life
Johannes Bergsten, Johan A. A. Nylander, Oscar E. Ospina, Alan R. Lemmon, Kelly B. Miller
Abstract
Diving beetles (family Dytiscidae) are important generalist predators in freshwater ecosystems that have been around since the Jurassic. Previous phylogenetic studies have identified a largely stable set of monophyletic named groups (subfamilies, tribes and subtribes); however, backbone relationships among these have remained elusive. Here we use whole genome sequencing to reconstruct the phylogeny of Dytiscidae. We mine de novo assemblies and combine them with others available from transcriptome studies of Adephaga to compile a dataset of 149 taxa and 5364 orthologous genes. Species tree and concatenated maximum likelihood methods provide largely congruent results, resolving in agreement all but two inter-subfamily nodes. All 11 subfamilies are monophyletic, supporting previous results; possibly also all tribes, but Hydroporini is recovered as paraphyletic with weak support and monophyly of Dytiscini is method dependent... Resolution of tribes in Dytiscinae is affected by methodological inconsistencies. Platynectini, new tribe, is described and Hydrotrupini redefined within subfamily Agabinae. This study is a step forward towards completely resolving the backbone phylogeny of Dytiscidae, which we hope will stimulate further work on remaining challenges.
Read the paper here:
https://resjournals.onlinelibrary.wiley.com/doi/10.1111/syen.12685
#beetle#diving beetle#dytiscidae#coleoptera#insect#entomology#taxonomy#cladistics#genetics#genomics#science#nature#animals
149 notes
·
View notes
Note
I’m a biology teacher, and we just finished our genetics chapter. I mentioned your blog to some of my students, and they asked if they could submit their own questions to your blog to see what animal they got. I’m so sorry if you have a random influx of 15 y/os in your inbox.
String identified: ’ a g tac, a t gtc cat. t g t tt, a t a t c t t t t g t at aa t gt. ’ a a a / .
Closest match: Patella vulgata genome assembly, chromosome: 1 Common name: Common European Limpet

(image source)
#tumblr genetics#genetics#biology#science#asks#anon#molluscs#sea snails#ocean#common european limpet#ohhhh ok that explains some things
651 notes
·
View notes
Text

Horses are among the world’s most elite athletes: When galloping, they can consume twice as much oxygen per kilogram as the fittest humans. All that oxygen supercharges horses’ cells’ energy-producing compartments as they crank out ATP, the chemical needed to power their impressive muscles. But making so much cellular fuel so quickly comes with a catch: the manufacture of pernicious byproduct molecules called reactive oxygen species (ROS), which can wreak havoc in cells.
How horses dealt with this biological trade-off and evolved into premier endurance athletes has long intrigued biologists. Researchers report today in Science that they have uncovered a big part of it, identifying a key mutation that lets horses safely produce so much ATP. The trait helped pave the way for horses to go from dog-size critters millions of years ago to the high-endurance athletes we know today.
The study’s detailed molecular work makes it “exceptional,” says José Calbet, an expert on the cellular responses to exercise at the University of Las Palmas de Gran Canaria who wasn’t involved with the study.
The mutation in question occurs in the gene that encodes a protein called KEAP1, which acts as a biochemical bouncer, binding to a different protein called NRF2 to prevent it from entering the cell’s nucleus, where it would otherwise activate stress-response genes that help blunt cell damage.
But ROS can help NRF2 sneak in by causing KEAP1 to release its bind on the protein, allowing it to enter the nucleus and trigger the cell’s stress-response genes.
Johns Hopkins University ophthalmologist and clinician scientist Elia Duh, a senior author of the new study, didn’t set out to study horses. Initially, Duh was interested in the KEAP1-NRF2 system because its role in activating stress-response genes makes it a tempting target for treating inflammation—and aging-related conditions, such as blinding retinal diseases, irritable bowel syndrome, and neurodegeneration.
Duh wondered whether any insights could be gleaned from studying the evolution of these proteins in different animals. So, he teamed up with Gianni Castiglione, an evolutionary biologist and biochemist at Vanderbilt University. Together, they scanned hundreds of vertebrate genomes looking for notable mutations to the gene for KEAP1.
The team’s genomic work revealed birds had almost completely lost the gene, presumably an adaptation to the extreme demands of flight. When they looked in horses, researchers noticed what initially appeared to be a DNA sequence that encoded an unusually short—and therefore presumably nonfunctional—version of the KEAP1 protein. But when Duh’s and Castiglione’s team grew horse cells in culture, it discovered the protein was very much there and working. “Naturally, I was worried I was doing something wrong,” Castiglione says. “Then one day, a light bulb went off.”
As it turns out, the computer algorithm scientists had used to scan the horse genome had made a mistake. The algorithm had spotted a specific kind of mutation in the part of the KEAP1 gene that changed the messenger RNA from CGA—which codes for the amino acid arginine—to UGA, which is what’s known as a “stop codon.”
Normally, the cellular machinery interprets UGA as a sign to stop translating the RNA into a protein. But instead, the horses’ genetic machinery recodes the stop codon into a different amino acid, cysteine, causing it to ignore that order. This phenomenon, known as a stop codon read-through, is common among viruses but rare in multicellular organisms.
“The identification of this evolutionarily significant UGA recoding event represents a potentially seminal finding, offering a model for uncovering other yet-unidentified cases of stop codon read-through,” says Hozumi Motohashi, a biologist at Tohoku University who has studied KEAP1 and NRF2.
That the replacement is a cysteine is particularly notable, Castiglione says. KEAP1 senses cellular stress through its cysteines, which contain sulfur atoms whose reactions with ROS, induce the chemical changes that cause KEAP1 to let go of NRF2. The mutation the researchers had identified adds another place on KEAP1 for ROS to interact, which makes the protein more sensitive to stress—and lets horse cells respond much faster to the cellular stress of intense exercise. “It does make complete sense [that] by introducing another cysteine, another sulfur, you would have heightened sensitivity,” Castiglione says.
What’s more, this tweaking of KEAP1 is a “[key] genetic component to the puzzle of the evolution of horses,” Duh says. “Once they figured out how to run, they could occupy all kinds of ecological niches,” Castiglione adds.
The finding could also point the way toward new kinds of drugs to treat diseases by targeting the specific parts of the KEAP1 protein that help horses hoof it. “By looking at what evolution has figured out, we know this is a viable strategy,” Castiglione says.
Source
834 notes
·
View notes
Text
‘Ferrari in a junkyard’: Mules sold at auction are rare, endangered horses

https://washingtonpost.com/climate-environment/2024/08/09/przewalskis-horses-rescued-dna-shrek-fiona/
Hannah Huckabay regularly combs livestock auctions online for horses she can rehabilitate and train at her Colorado ranch. But when she saw a video in February of a mule for sale in Kansas, she could hardly believe what she was seeing.
The stocky animal’s short black mane shot straight up like a mohawk, and its white belly stood out against its tan coat. As it nervously paced in its corral, Huckabay said it bore a striking resemblance to Przewalski’s horse, a critically endangered species she’d learned about while studying equine science.
“I was like, ‘There is no way. That is not a mule,’” Huckabay recalled thinking. “That’s a purebred Przewalski.”
Such a find would be incredibly rare. Once extinct in the wild, around 2,500 Przewalski’s horses remained worldwide as of 2022. They’re native to Mongolia and in June, seven were reintroduced to nearby Kazakhstan as part of an effort to return them to their natural habitats. They are the only truly wild horse remaining (mustangs are feral horses).
But scientists say Huckabay’s hunch appears to be correct. Hair samples from the animal Huckabay purchased - along with a second horse recently surrendered at a Utah sanctuary - were sent to Texas A&M University’s animal genetics lab. Both appear to be Przewalski’s horses, said Rytis Juras, the genetics lab’s director who tested both samples.
The hair test looks for genetic markers associated with different horse breeds to determine an animal’s likely ancestry. Unequivocally confirming that the horses are purebred Przewalski’s and not hybrids would require advanced blood tests that are expensive and would mean sedating the equines.
The blood tests look at the number of chromosomes in a horse’s cells - 66 in a purebred Przewalski, versus 64 in a common horse or 62 in a donkey. An even more advanced version could sequence the horse’s entire genome.
But Juras and two other scientists who reviewed the findings said the hair-test results are reliable.
“If I would have gotten it from a zoo … that would be one thing,” Juras said of receiving the samples. But two random tests with Przewalski’s results were “surprising and a little bit disturbing,” he said. “This is weird.”
How the horse Huckabay found - and the second in Utah - ended up in livestock auctions is a mystery, said Christopher Faulk, a professor of animal science at the University of Minnesota who has studied Przewalski’s horse genetics and also reviewed the DNA results.
“Someone had to have known what they were, they don’t just appear out of anywhere,” Faulk told The Post. “Especially to have been disposed of in that way is even weirder,” he said, since livestock that aren’t purchased at auction can end up in slaughterhouses.
“That’s like finding a Ferrari in a junkyard,” he added.
Huckabay bought the animal for $1,375 in February and, after three weeks in quarantine, the ragged and underweight animal sold as a mule arrived at her ranch outside Denver.
Seeing its features in-person left her even more convinced it was a Przewalski’s horse, she said. With a large clunky head and stiff black mane, her daughter said the horse was so ugly, he was cute, Huckabay recalled. They named him Shrek, after DreamWorks’s beloved ogre.
After almost two months of helping Shrek acclimate, Huckabay’s daughter stumbled upon a video posted on June 9 from a sanctuary in Utah.
“Did we just have a Przewalski mare surrendered?!” the caption read.
Kelsey and Gunnar Bjorklund - who own the Lazy B Equine Rescue and Sanctuary in Utah - suspected their mare was also a Przewalski. But they had no idea there was a second possible Przewalski, saved from another auction.
The Bjorklunds’ horse was brought to their facility after being purchased for $35 in January at an auction in Utah, where she was advertised as a mule.
“It takes more money to get your nails done,” Kelsey said, adding that her previous owner decided to surrender the mare after she flunked out of a professional training program.
When the horse arrived and was unloaded from the trailer, “we were just in shock,” Gunnar said. It was clear the animal wasn’t a mule or a mustang, he said.
“Anyone getting possible Przewalski vibes!?” the Bjorklunds posted. “A true wild, endangered species of equine‼️ How cool would that be!”
In response to seeing the Bjorklunds’ viral video, Huckabay’s daughter posted her own videos of Shrek two days later. One got over 11 million views.
After coming across Shrek’s video, it was easy for the Bjorklunds to settle on a name for their mystery horse - Fiona, the princess-heroine from the Shrek movies.
The rescuers were stunned that two possible Przewalski’s horses could have surfaced almost simultaneously. The Endangered Species Act allows private ownership of endangered animals, but only with a permit, and under strict stipulations. The law prohibits the possession of illegally obtained endangered animals or their transport across state lines without permits.
The U.S. Fish and Wildlife Service declined to comment on whether officials are investigating the horses’ chain of custody.
Some livestock auctions have occasionally served as hubs for illicit trade in exotic animal species.
Because most Przewalski’s horses descend from only about a dozen surviving individuals, scientists closely manage breeding genetics for diversity. Compared to the feral mustang, Przewalski’s are more resilient, said Dolores Reed, a biologist who helps oversee a small herd of the endangered horses at the Smithsonian’s National Zoo and Conservation Biology Institute. Przewalski’s horses are built for the Mongolian steppe’s harsh climate, she said, adding, “they’re very tough,” and can be unpredictable.
There are about 100 Przewalski’s horses in U.S. zoos, Reed said.
Shrek and Fiona are adjusting to their new environments, their owners said. After keeping his distance from people and trotting in circles in his pen while stressed, Shrek has relaxed and moved to a larger field. He has bonded with two gentle mares and while he won’t accept treats from people’s hands, he loves when apples and carrots are left in his feed bucket, Huckabay said.
“He’s very piggy,” she said.
In Utah, Fiona has put on weight and made friends with a miniature mule and a quarter horse filly at the Bjorklunds’ sanctuary.
The rescuers wonder what would’ve happened if Shrek and Fiona hadn’t been saved. The endangered animals might’ve been sent to slaughter “and nobody would have known about it,” Gunnar said.
Huckabay and the Bjorklunds plan to care for the horses as long as needed, but said they’d prefer to see their rescued Przewalski’s move to a professional conservation program.
Shrek is happy on the ranch, but Huckabay said she’d rather see him with “a herd of his own.”
“That would be the best-case scenario,” she said.
#this is fucking insane#Przewalski’s horses#Przewalski’s horse#horses#colorado#animal protection#animal welfare#science#environment#nature#animals#usa#long post
1K notes
·
View notes
Note
D’you perchance have any thoughts on the morphological (for lack of a better word?) dire wolves that Colossal Biosciences just revealed to the public? 👀

Oh my god Aenocyon, you can't just ask someone why they're white!
"Morphological dire wolf" my ass. Which is coincidentally where Colossal pulled the white coats from…
Give me an example of a modern temperate/grassland predator that's white*, I'll wait. *Excluding white lions, which are an uncommon but resilient morph resulting from leucism.
I based my Aenocyon design off bushdogs and dholes. They are called Masked Wolves in Kindred's setting, because I enjoy a good pseudo hyena niche uvu-b
Extremely extremely long 'thoughts' below the cut lol c':
Preface: in this discussion the term "dire wolf" has too many meanings, as such I will be referring to them as follows:
Thrones' wolves: for the huge, white, fantasy animals from Game Of Thrones GMO wolves: for Romulus, Remus and Khaleesi, Colossal's creations, Canis lupus Aenocyon: for Aenocyon dirus, the true, extinct dire wolf known from fossils across North America
----
Part 1: That's not a dire wolf-
The first question everyone has been asking is "So, are dire wolves de extinct now?" The answer is an emphatic "NO!" from anyone with knowledge of genetics, palaeontology, or taxonomy.
Aenocyon dirus were actually not wolves, nor dogs, but a secret third thing.
They are canids, but last shared a common ancestor with grey wolves and their lineage some ~5.7 million years ago.
For context, this paper suggests a similar divergence time between genus Homo (humans, Neanderthals and co) and Pan (chimps and bonobos); animals that look and behave markedly differently from each other.
The genomes of Canis lupus and Aenocyon dirus being 99.5% similar may sound like a lot, but again, humans share 98.8% with chimps, and 99.7% with Neanderthals, and yet are very distinct from both.
Skeletally, behaviourally, in soft tissue, etc, you could tell any of the three apart; the same goes for Aenocyon and Canis members.
Additionally, Colossal made 20 changes in 14 genes.
The grey wolf genome has 2,447,000,000 base pairs. Does that maths seem a bit off to you?
That's not even enough to change a grey wolf into a domestic dog, let alone an ancient outgroup!
This would be akin to modifying a lion to have bigger teeth and saying you resurrected Smilodon fatalis.
Or editing a Asian Elephant genome so they retain their juvenile hair and calling it a Woolly Mammoth.
It's a bold-faced lie.
Beth Shapiro says "they look and act like dire wolves" but that, too,simply isn't true.
Visually, the GMO wolves simply aren't what Aenocyon would have looked like. It's what a Thrones' wolf looks like.
Hmmmmm, funny about that, seeing George R R Martin helped fund the 'dire wolf project'...
As with many fossil animals, we don't know much about Aenocyon's behaviour.
You can't say the GMO wolves (who are also still pups) act like Aenocyon, because that's based off nothing.
What we do know is Aenocyon were likely pack animals (from the sheer number found in La Brea Tarpits), and crunched more bones than modern wolves (from their many broken teeth).
Also, crucially, they had Wild Sex Lives (from the many, huge, broken and healed bacula... youch).
Colossal is also being colossally shady by: doubling down on their bs use of the outdated "morphological species definition", blatantly misleading the public with their use of the words 'cloning', 'dire wolves', and 'de extinction', and refusing to share their methods in a peer reviewed paper before going public with a clickbait headline.
Do not trust them with your Red wolves either. They're using coyote hybrids and considering what they deem 'close enough' for a dire wolf, I wouldn't put any money on the quality of their GMO red wolves either...
Also can I just say, whatever genes they modified to "make the skull larger" clearly didn't impact the lower jaw...

No, I'm not sorry for this image uvu-b (But for real look at that poor pup and his overbite jfc)
Part 2: -and if it was, that wouldn't be good either.
I fundamentally do not support de extinction.
No, not even for the Thylacine, not even for passenger pigeons, nor the dodo. Even my beloved Homotherium should be left in the past.
This might be an unexpected stance because I am, surprising no one, a big fan of extinct animals, megafauna and otherwise.
But the thing is, I'm an even bigger fan of actual, living animals.
The animal ethics of de extinction are dubious at best.
The surrogate dog mothers of the GMO wolves likely won't live good lives.
I wouldn't be surprised if they were destroyed after being used, because their bodies could contain feto microchimerisms and Colossal absolutely doesn't want their special wolf genome getting out.
I doubt the GMO wolves themselves will live a full life before they outgrow their hearts, like Ligers.
This would likely be the case for any modern animal genetically modified into megafauna; a body not adapted to deal with the increased size.
Purely conjecture, but I also wouldn't be surprised if Romulus, Remus and Khaleesi have vision/hearing issues from their white coats.
White coats in wolves are associated with hearing impairments, so the gene used for these animals was from domestic dogs. Meaning Colossal has created a very expensive wolfdog.
Again, what kind of life are these wolfdogs supposed to live? As awful pets for the rich? In a zoo? Released to pollute wild wolf genomes? (assuming they're fertile; I hope not)
Regardless, it's not looking good if they ever planned to have them be 'wild animals'
Even true clones (which the GMO wolves are not) tend to have health issues.
Celia the Pyrenean Ibex (bucardo) was cloned, but the clone died after 9 minutes from a deformed lung.
So in 2003, this made the bucardo the first species to go extinct twice, yippee?
There's also the problem of genetic diversity.
How many intact genomes do you have on hand?
For dire wolves the answer is Zero!
To my knowledge, we don't have the full genome coded from one individual, just Frankenstein-ed from many. Which is fine for sequencing the canine family tree's relatedness, but not for cloning.
The absolute minimum individuals to survive a genetic bottleneck is said to be 50 in larger species. Called the 50/500 rule, it states that 50 is enough to survive, but 500 is required to prevent genetic drift.
To which I say, good luck!
Even with well preserved permafrost species (such as woolly mammoths), you'll have a hard time finding 500 individuals with prefect genomes.
And then, where will you put them?
If you were to, somehow, make a breeding population, where are they going? A national park? A zoo? Is their old habitat still available to them?
In Aenocyon, the answer is simply "they don't have a niche anymore".
Unlike the Thylacine or Dodo, humans did not directly cause the extinction of Aenocyon dirus. And even if they had, it was 10,000 years ago!
Would making room for a de extinct species impact the habitat/niche of another species?
Regular grey wolves fill Aenocyon's role as a canine mesopredator, with Puma as the apex (alongside bears as an apex omnivore).
With the loss of megafauna to prey on, a de extinct predator would just compete with other, also endangered species.
Animals also change the environment they life in.
Mammoths will clear trees like modern elephants. This would recreate the Mammoth Steppe, but those trees making up the taiga and boreal forests are themselves crucial habitat.
Other species have moved in since the mammoths' extinction. Siberian tigers, lynx, muskoxen, brown bears, elk, moose, and so many others; many endangered.
Trees also prevent erosion, which is already happening at unprecedented rates due to agriculture and deforestation.
Crucially: What's to stop an extinct animal going the same way it went out last time?
Ask yourself this:
Would the average American appreciate "flocks of Passenger pigeons big enough to darken the sky and whiten ground with their guano"?
Would people suddenly be okay with lions in Europe eating their livestock, when they are champing the bit to shoot Iberian wolves again?
Would Tasmanians suddenly feel the same about the Thylacine, when farmers in Australia still happily kill dingoes and eagles for lamb predation? [citation, I am an enviro technician and have had farmers tell me they shoot Wedge-tails, knowing I'm a toothless lion to stop them.]
I doubt it
At what cost?
Are we going to find 50 thylacine genomes?
If so (doubtful), how much will cloning and/or modifying a relative into a thylacine cost? Now that x50?
Wouldn't that money be better spent on quoll reintroduction?
What about finding 50 gestational carriers for mammoths?
Are you going to use their closest relative; the already critically endangered Asian Elephant?
Wouldn't that time and effort on those elephant mothers be better used making more elephants?
And the social cost:
If extinction isn't forever, what's to incentivize lawmakers to fund conservation?
Really, it comes down to this:
Why bring back the dire wolf when we could put this money into protecting the Iberian and Red wolves?
Why bring back the thylacine when their cousin is dying of a transmissible cancer?
We've already seen the impacts of "extinction isn't forever anymore", with those in power already trying to cut funding to conservation, because you can "just bring them back".
But as we've seen time and time again: there is no Planet B. There is no De-Extinction, not really.
Maybe what was gone should stay gone, so we can focus on what we still have.
#*farkin mike drop*#whoops this took an extremely long time I can't be trusted not to write a thesis for things like this bc im Passionate#sorry not sorry for the colours- it makes it easier for my brain so I hope it helps this site full of other ND people lolol#also ur getting this instead of a Kindred update bc i have not been able to work on pages there's been 6767687 family members here all week#mammothask#stressingcosmos#GMO wolves#<- my tag for these poor beasts#bc they sure aren't dire wolves#bc u see dire wolves are#aenocyon#dire wolf#masked wolf#romulus remus and khaleesi#de extinction#animal ethics#scientific ethics#paleo stuff#sorta#wolf#grey wolf#gray wolf#pavlova pictures#bc i drew this
380 notes
·
View notes