Text
Week 4
INPUT
import pandas import numpy import seaborn import matplotlib.pyplot as plt # any additional libraries would be imported here
mydata = pandas.read_csv('prostate_1.csv', low_memory=False)
# subset variables in new data frame, sub1 sub1=mydata[['AgeF','PSA', 'Cancer Volume']]
a = sub1.head print(a)
#new PSA variable, categorical 1 through 2 def PSA (row): if row['PSA'] < 4 : return 1 if row['PSA'] > 4 : return 2 sub1['PSA'] = sub1.apply (lambda row: PSA (row),axis=1)
a = sub1.head print(a)
#new Age variable, categorical 1 through 2 def Age (row): if row['AgeF']== '41-50': return 1 if row['AgeF'] == '51-60' : return 2 if row['AgeF'] == '61-70' : return 3 if row['AgeF'] == '71-80' : return 4 sub1['Age'] = sub1.apply (lambda row: Age (row),axis=1)
a = sub1.head print(a)
#new Cancer_Volume variable, categorical 1 through 4 def Cancer_Volume(row): if row['Cancer Volume'] < 11: return 1 if row['Cancer Volume'] >11 and row['Cancer Volume'] < 21 : return 2 if row['Cancer Volume'] >21 and row['Cancer Volume'] <31 : return 3 if row['Cancer Volume'] >31: return 4 sub1['Cancer_Volume'] = sub1.apply (lambda row: Cancer_Volume(row),axis=1)
a = sub1.head print(a)
#univariate bar graph for categorical variables for PSA level # First hange format from numeric to categorical sub1["PSA"] = sub1["PSA"].astype('category')
seaborn.countplot(x="PSA", data=sub1) plt.xlabel('PSA level') plt.title('PSA level Among Adult men who visited the university medical center in the Prostate cancer Study')
#univariate bar graph for categorical variables for Age groups # First hange format from numeric to categorical sub1["AgeF"] = sub1["AgeF"].astype('category')
seaborn.countplot(x="AgeF", data=sub1) plt.xlabel('AgeF') plt.title('Age groups Among Adult men who visited the university medical center in the Prostate cancer Study')
#univariate bar graph for categorical variables for Cancer Volume # First hange format from numeric to categorical sub1["Cancer_Volume"] = sub1["Cancer_Volume"].astype('category')
seaborn.countplot(x="Cancer_Volume", data=sub1) plt.xlabel('Cancer_Volume') plt.title('Cancer Volume Among Adult men who visited the university medical center in the Prostate cancer Study')
# standard deviation and other descriptive statistics for quantitative variables
print ('PSA level') desc2 = sub1['PSA'].describe() print (desc2)
c1= sub1.groupby('PSA').size() print (c1)
print ('mode PSA level') mode1 = sub1['PSA'].mode() print (mode1)
c1= sub1.groupby('PSA').size() print (c1)
p1 = sub1.groupby('PSA').size() * 100 / len(mydata) print (p1)
# standard deviation and other descriptive statistics for quantitative variables
print ('Age') desc2 = sub1['AgeF'].describe() print (desc2)
c2= sub1.groupby('AgeF').size() print (c2)
print ('mode of Age') mode1 = sub1['AgeF'].mode() print (mode1)
p2 = sub1.groupby('AgeF').size() * 100 / len(mydata) print (p2)
print ('Cancer Volume') desc2 = sub1['Cancer_Volume'].describe() print (desc2)
c2= sub1.groupby('Cancer_Volume').size() print (c2)
print ('Mode of Cancer Volume') mode1 = sub1['Cancer_Volume'].mode() print (mode1)
# bivariate bar graph C->Q seaborn.factorplot(x='AgeF', y='PSA', data=mydata, kind="bar", ci=None) plt.xlabel('Age') plt.ylabel('PSA level')
OUTPUT
<bound method NDFrame.head of AgeF PSA Cancer Volume 0 41-50 0.651 0.5599 1 51-60 0.852 0.3716 2 71-80 0.852 0.6005 3 51-60 0.852 0.3012 4 61-70 1.448 2.1170 .. ... ... ... 92 61-70 80.640 16.9455 93 41-50 107.770 45.6042 94 51-60 170.716 18.3568 95 61-70 239.847 17.8143 96 61-70 265.072 32.1367
[97 rows x 3 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume 0 41-50 1 0.5599 1 51-60 1 0.3716 2 71-80 1 0.6005 3 51-60 1 0.3012 4 61-70 1 2.1170 .. ... ... ... 92 61-70 2 16.9455 93 41-50 2 45.6042 94 51-60 2 18.3568 95 61-70 2 17.8143 96 61-70 2 32.1367
[97 rows x 3 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume Age 0 41-50 1 0.5599 1 1 51-60 1 0.3716 2 2 71-80 1 0.6005 4 3 51-60 1 0.3012 2 4 61-70 1 2.1170 3 .. ... ... ... ... 92 61-70 2 16.9455 3 93 41-50 2 45.6042 1 94 51-60 2 18.3568 2 95 61-70 2 17.8143 3 96 61-70 2 32.1367 3
[97 rows x 4 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume Age Cancer_Volume 0 41-50 1 0.5599 1 1 1 51-60 1 0.3716 2 1 2 71-80 1 0.6005 4 1 3 51-60 1 0.3012 2 1 4 61-70 1 2.1170 3 1 .. ... ... ... ... ... 92 61-70 2 16.9455 3 2 93 41-50 2 45.6042 1 4 94 51-60 2 18.3568 2 2 95 61-70 2 17.8143 3 2 96 61-70 2 32.1367 3 4
[97 rows x 5 columns]> PSA level count 97 unique 2 top 2 freq 83 Name: PSA, dtype: int64 PSA 1 14 2 83 dtype: int64 mode PSA level 0 2 Name: PSA, dtype: category Categories (2, int64): [1, 2] PSA 1 14 2 83 dtype: int64 PSA 1 14.43299 2 85.56701 dtype: float64 Age count 97 unique 4 top 61-70 freq 59 Name: AgeF, dtype: object AgeF 41-50 8 51-60 17 61-70 59 71-80 13 dtype: int64 mode of Age 0 61-70 Name: AgeF, dtype: category Categories (4, object): [41-50, 51-60, 61-70, 71-80] AgeF 41-50 8.247423 51-60 17.525773 61-70 60.824742 71-80 13.402062 dtype: float64 Cancer Volume count 97 unique 4 top 1 freq 75 Name: Cancer_Volume, dtype: int64 Cancer_Volume 1 75 2 16 3 4 4 2 dtype: int64 Mode of Cancer Volume 0 1 Name: Cancer_Volume, dtype: category Categories (4, int64): [1, 2, 3, 4]
The univariate graph of PSA level:
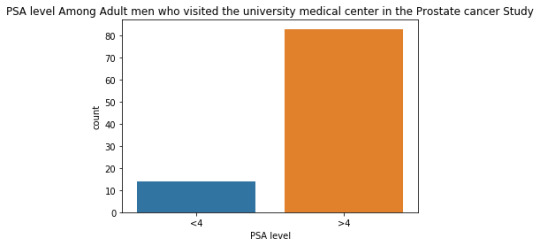
This graph is unimodal, with its highest peak at the�� category of >4 PSA level . It seems to be skewed to the left as there are higher frequencies in higher category(>4) than the lower category.
The univariate graph of Age groups:
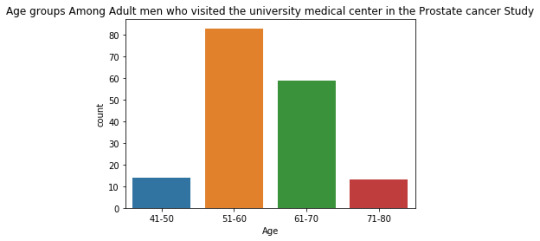
This graph is unimodal, with its highest peak at 51 to 60 age group. It seems to be skewed to the right as there are higher frequencies in the lower age ranges from 51 to 60.
The univariate graph of Cancer Volume:
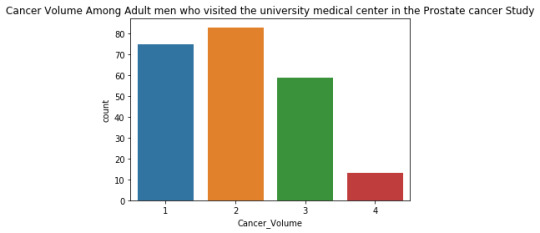
This graph is unimodal, with its highest peak at the category of 2 (11-20) . It seems to be skewed to the right as there are higher frequencies in lower categories than the higher categories.

The graph above plots the Cancer Volume of the adult men to the adult men corresponding Age groups. We can see that the bar chat does not show a clear relationship/trend between the two variables.
0 notes
Text
Week 3
import pandas
import numpy
# any additional libraries would be imported here
mydata = pandas.read_csv('prostate_1.csv', low_memory=False)
# subset variables in new data frame, sub1
sub1=mydata[['AgeF','PSA', 'Cancer Volume']]
a = sub1.head
print(a)
#new PSA variable, categorical 1 through 2
def PSA (row):
if row['PSA'] < 4 :
return 1
if row['PSA'] > 4 :
return 2
sub1['PSA'] = sub1.apply (lambda row: PSA (row),axis=1)
a = sub1.head
print(a)
#new Age variable, categorical 1 through 2
def Age (row):
if row['AgeF']== '41-50':
return 1
if row['AgeF'] == '51-60' :
return 2
if row['AgeF'] == '61-70' :
return 3
if row['AgeF'] == '71-80' :
return 4
sub1['Age'] = sub1.apply (lambda row: Age (row),axis=1)
a = sub1.head
print(a)
#new Cancer_Volume variable, categorical 1 through 4
def Cancer_Volume(row):
if row['Cancer Volume'] < 11:
return 1
if row['Cancer Volume'] >11 and row['Cancer Volume'] < 21 :
return 2
if row['Cancer Volume'] >21 and row['Cancer Volume'] <31 :
return 3
if row['Cancer Volume'] >31:
return 4
sub1['Cancer_Volume'] = sub1.apply (lambda row: Cancer_Volume(row),axis=1)
a = sub1.head
print(a)
#frequency distributions for primary and secondary ethinciity variables
print( 'counts for PSA level')
c10 = sub1['PSA'].value_counts(sort=False)
print(c10)
print( 'percentages for PSA level')
p10 = sub1['PSA'].value_counts(sort=False, normalize=True)
print (p10)
print('counts for Age')
c11 = sub1['Age'].value_counts(sort=False)
print(c11)
print( 'percentages for Age')
p11= sub1['Age'].value_counts(sort=False, normalize=True)
print (p11)
print( 'counts for Cancer Volume')
c12 = sub1['Cancer_Volume'].value_counts(sort=False)
print(c12)
print( 'percentages for Cancer Volume')
p12 = sub1['Cancer_Volume'].value_counts(sort=False, normalize=True)
print (p12)
Output
runfile('C:/Users/NAOMI/Downloads/Documents/Cousera/Data Visualization/week 3/Assignment 3 new.py', wdir='C:/Users/NAOMI/Downloads/Documents/Cousera/Data Visualization/week 3') <bound method NDFrame.head of AgeF PSA Cancer Volume 0 41-50 0.651 0.5599 1 51-60 0.852 0.3716 2 71-80 0.852 0.6005 3 51-60 0.852 0.3012 4 61-70 1.448 2.1170 .. ... ... ... 92 61-70 80.640 16.9455 93 41-50 107.770 45.6042 94 51-60 170.716 18.3568 95 61-70 239.847 17.8143 96 61-70 265.072 32.1367
[97 rows x 3 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume 0 41-50 1 0.5599 1 51-60 1 0.3716 2 71-80 1 0.6005 3 51-60 1 0.3012 4 61-70 1 2.1170 .. ... ... ... 92 61-70 2 16.9455 93 41-50 2 45.6042 94 51-60 2 18.3568 95 61-70 2 17.8143 96 61-70 2 32.1367
[97 rows x 3 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume Age 0 41-50 1 0.5599 1 1 51-60 1 0.3716 2 2 71-80 1 0.6005 4 3 51-60 1 0.3012 2 4 61-70 1 2.1170 3 .. ... ... ... ... 92 61-70 2 16.9455 3 93 41-50 2 45.6042 1 94 51-60 2 18.3568 2 95 61-70 2 17.8143 3 96 61-70 2 32.1367 3
[97 rows x 4 columns]> <bound method NDFrame.head of AgeF PSA Cancer Volume Age Cancer_Volume 0 41-50 1 0.5599 1 1 1 51-60 1 0.3716 2 1 2 71-80 1 0.6005 4 1 3 51-60 1 0.3012 2 1 4 61-70 1 2.1170 3 1 .. ... ... ... ... ... 92 61-70 2 16.9455 3 2 93 41-50 2 45.6042 1 4 94 51-60 2 18.3568 2 2 95 61-70 2 17.8143 3 2 96 61-70 2 32.1367 3 4
[97 rows x 5 columns]> counts for PSA level 1 14 2 83 Name: PSA, dtype: int64 percentages for PSA level 1 0.14433 2 0.85567 Name: PSA, dtype: float64 counts for Age 1 8 2 17 3 59 4 13 Name: Age, dtype: int64 percentages for Age 1 0.082474 2 0.175258 3 0.608247 4 0.134021 Name: Age, dtype: float64 counts for Cancer Volume 1 75 2 16 3 4 4 2 Name: Cancer_Volume, dtype: int64 percentages for Cancer Volume 1 0.773196 2 0.164948 3 0.041237 4 0.020619 Name: Cancer_Volume, dtype: float64
I created new data with three variables: AgeF, PSA and Cancer Volume. The were no missing data set in my data. For Age, the most commonly endorsed is 3 (60.8%) , meaning more than half of the men who went for the checkup are from the age 61-70 years. For PSA, 2 (85.57%) has the highest percentage, meaning the PSA which is greater than 4 has the highest frequency of 83. For Cancer Volume, 1 ( 77.32% ) has the highest percentage among the others which means the Cancer Volume less than 10 has the highest frequency of 75 with 77.32%.
0 notes
Text
My program (code)
import pandas import numpy
mydata = pandas.read_csv("prostate_1.csv", low_memory = False)
print(len(mydata)) #prints out number of observations (row) print(len(mydata.columns)) #prints out number of columns(variables)
mydata["Age"]= mydata["AgeF"] mydata["PSA"]= mydata["PSAF"]
#counts and percentages (i.e. frequency distributions) for each variable print("count for Age groups") count1 = mydata['Age'].value_counts(sort=False) print (count1)
print("percentages for Age groups") p1 = mydata['Age'].value_counts(sort=False, normalize=True) print (p1)
print("count for PSA level") #PSA was grouped into 2 that is <4 and >4 count2 = mydata["PSA"].value_counts(sort=False) print(count2)
print("percentages for PSA level ") p2 = mydata["PSA"].value_counts(sort=False, normalize=True) print(p2)
print("count for Weight of cancer in gm") count4 = mydata["Weight"].value_counts(sort=False) print(count4)
print("percentages for Weight of cancer in gm ") p4 = mydata["Weight"].value_counts(sort=False, normalize = True) print(p4)
# freqeuncy disributions using the 'bygroup' function cot1= mydata.groupby('Age').size() print(cot1)
pot1 = mydata.groupby('Age').size() * 100 / len(mydata) print(pot1)
cot2= mydata.groupby('PSA').size() print(cot2)
pot2 = mydata.groupby('PSA').size() * 100 / len(mydata) print (pot2)
cot4= mydata.groupby('Weight').size() print (cot4)
pot4 = mydata.groupby('Weight').size() * 100 / len(mydata) print (pot4)
#upper-case all DataFrame column names - place afer code for loading data aboave mydata.columns = map(str.upper, mydata.columns)
# bug fix for display formats to avoid run time errors - put after code for loading data above pandas.set_option('display.float_format', lambda x:'%f'%x)
Output
97
12
count for Age group
51-60 17
41-50 8
71-80 13
61-70 59
Name: Age, dtype: int64
percentages for Age groups
51-60 0.175258
41-50 0.082474
71-80 0.134021
61-70 0.608247
Name: Age, dtype: float64
count for PSA level
>4 83
<4 14
Name: PSA, dtype: int64
percentages for PSA level
>4 0.855670
<4 0.144330
Name: PSA, dtype: float64
count for Weight
31.500000 1
29.964000 1
10.697000 1
21.542000 1
59.740000 2
..
23.104000 1
22.646000 1
45.604000 1
39.646000 2
42.948000 1
Name: Weight, Length: 77, dtype: int64
percentages for Weight
31.500000 0.010309
29.964000 0.010309
10.697000 0.010309
21.542000 0.010309
59.740000 0.020619
23.104000 0.010309
22.646000 0.010309
45.604000 0.010309
39.646000 0.020619
42.948000 0.010309
Name: Weight, Length: 77, dtype: float64
Age
41-50 8
51-60 17
61-70 59
71-80 13
dtype: int64
Age
41-50 8.247423
51-60 17.525773
61-70 60.824742
71-80 13.402062
dtype: float64
PSA
<4 14
>4 83
dtype: int64
PSA
<4 14.432990
>4 85.567010
dtype: float64
Weight
10.697000 1
14.732000 1
15.959000 1
17.637000 1
20.086000 1
..
83.931000 1
91.836000 1
112.168000 1
119.104000 1
450.339000 1
Length: 77, dtype: int64
Weight
10.697000 1.030928
14.732000 1.030928
15.959000 1.030928
17.637000 1.030928
20.086000 1.030928
83.931000 1.030928
91.836000 1.030928
112.168000 1.030928
119.104000 1.030928
450.339000 1.030928
Length: 77, dtype: float64
runfile('C:/Users/NAOMI/Documents/Anaconda/Anaconda work/Assignment.py', wdir='C:/Users/NAOMI/Documents/Anaconda/Anaconda work')
97
12
count for Age group
51-60 17
41-50 8
71-80 13
61-70 59
Name: Age, dtype: int64
percentages for Age groups
51-60 0.175258
41-50 0.082474
71-80 0.134021
61-70 0.608247
Name: Age, dtype: float64
count for PSA level
>4 83
<4 14
Name: PSA, dtype: int64
percentages for PSA level
>4 0.855670
<4 0.144330
Name: PSA, dtype: float64
count for Weight
31.500000 1
29.964000 1
10.697000 1
21.542000 1
59.740000 2
..
23.104000 1
22.646000 1
45.604000 1
39.646000 2
42.948000 1
Name: Weight, Length: 77, dtype: int64
percentages for Weight
31.500000 0.010309
29.964000 0.010309
10.697000 0.010309
21.542000 0.010309
59.740000 0.020619
23.104000 0.010309
22.646000 0.010309
45.604000 0.010309
39.646000 0.020619
42.948000 0.010309
Name: Weight, Length: 77, dtype: float64
Age
41-50 8
51-60 17
61-70 59
71-80 13
dtype: int64
Age
41-50 8.247423
51-60 17.525773
61-70 60.824742
71-80 13.402062
dtype: float64
PSA
<4 14
>4 83
dtype: int64
PSA
<4 14.432990
>4 85.567010
dtype: float64
Weight
10.697000 1
14.732000 1
15.959000 1
17.637000 1
20.086000 1
..
83.931000 1
91.836000 1
112.168000 1
119.104000 1
450.339000 1
Length: 77, dtype: int64
Weight
10.697000 1.030928
14.732000 1.030928
15.959000 1.030928
17.637000 1.030928
20.086000 1.030928
83.931000 1.030928
91.836000 1.030928
112.168000 1.030928
119.104000 1.030928
450.339000 1.030928
Length: 77, dtype: float64
This result shows that the adult from age 61-70 has the highest frequency with 61% followed by 51-60. The PSA level was grouped into two under <4 and >4. The PSA level with >4 has the highest frequency with 86%.
0 notes
Text
Regression Analysis of the factors affecting high grade prostate cancer in patients.
I have decided that I am particularly interested in high grade prostate cancer and this my own data set. I have included all the factors which I think are the main causes of high grade prostate cancer. Examples of factors include age of patients, prostate weight.
Research Questions
1. Does age positively affect high grade prostate cancer?
2. Is there a high correlation between high grade prostate cancer and the various factors?
Topic of Interest
Regression Analysis of the factors affecting high grade prostate cancer in patients.
Second (Related) Topic of Interest
Correlation among variables stated in our codebook.
Correlation is the measure of the strength of the linear relationship among, usually, continuous random variables. Correlation is always between -1 and +1. Values closer to -1 and +1 indicate high negative and high positive correlation respectively i.e. a strong positive or negative association. Correlation values closer to zero indicates weak relationship among the variables of interest.
Literature Review
Prostate cancer is the second most common diagnosed cancer and the fourth leading cause of cancer death in men worldwide (WCRF 2019). Physicians use rectal examination and prostate-specific antigen (PSA) concentration in blood to detect prostate cancer (Catalona et al. 1997, Heindenreich et al. 2014), the former is not welcome because of psychological implications and the latter could yield false-positive or false-negative results.
The severity of prostate cancer and survival probability of diagnosed patients can be estimated with the Gleason Score (Stark et al. 2009), but its accuracy and precision depend on multiple biopsies (PCEC 2019), another invasive and traumatic method. Other variables that might be associated to Gleason Score that can be obtained by less invasive methods and may be used to predict prostate cancer risk, for instance: prostate weight, benign prostate hyperplasia and seminal vesicle invasion, can be effectively detected and measured using ultrasonography (Kilic et al. 2014, Soylu et al. 2013).
The use of morphological and physiological parameters measured by noninvasive methods to predict the presence of high-grade prostate cancer remains unstudied. In this work we used seven morphological and physiological variables that can be measured in blood samples and ultrasonography to estimate the probability of presence of high-grade prostate cancer and therefore reduced the psychological impact of invasive diagnostic methods. It was recognized that age was the predominant factor affecting high grade prostate cancer (Naveda et al. 2019).
Hypothesis
Age is the principal determinant of high grade cancer. The probability of presence of high-grade prostate cancer increases as a person ages.
CodeBook
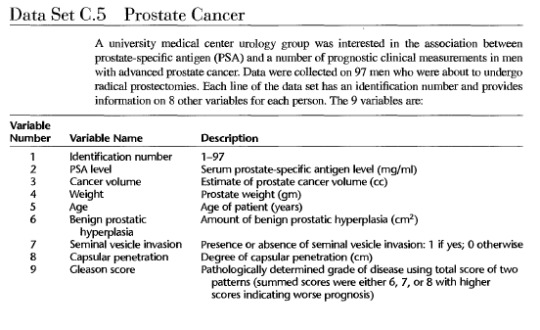
Adapted in part from: Hastie, T. J.; R. J. Tibshirani; and J. Friedman. The Elements of Statistical Learning: Data Mining. Inference. And Prediction. New York: Springer-Verlag, 2001.
Applied Linear Regression Models edition 5 Kutner et al.
1 note
·
View note