#Agef 21
Text
Le Chemin Gourmand de Nuits-Saint-Georges 2022 fait le plein
Le Chemin Gourmand de Nuits-Saint-Georges 2022 fait le plein
La 10e balade gourmande de Nuits-Saint-Georges affiche complet trois mois avant l’événement du 26 juin 2022. Son papa Pierre Mostacci est naturellement ravi.
Au taquet ! Le Chemin Gourmand de Nuits-Saint-Georges 2022 accueillera 1800 participants. © D.R.
Au bout du fil, Pierre Mostacci a le sourire. Il en serait presque gêné au moment d’annoncer que le Chemin Gourmand de Nuits-Saint-Georges…

View On WordPress
0 notes
Text
Week 4
INPUT
import pandas
import numpy
import seaborn
import matplotlib.pyplot as plt
# any additional libraries would be imported here
mydata = pandas.read_csv('prostate_1.csv', low_memory=False)
# subset variables in new data frame, sub1
sub1=mydata[['AgeF','PSA', 'Cancer Volume']]
a = sub1.head
print(a)
#new PSA variable, categorical 1 through 2
def PSA (row):
if row['PSA'] < 4 :
return 1
if row['PSA'] > 4 :
return 2
sub1['PSA'] = sub1.apply (lambda row: PSA (row),axis=1)
a = sub1.head
print(a)
#new Age variable, categorical 1 through 2
def Age (row):
if row['AgeF']== '41-50':
return 1
if row['AgeF'] == '51-60' :
return 2
if row['AgeF'] == '61-70' :
return 3
if row['AgeF'] == '71-80' :
return 4
sub1['Age'] = sub1.apply (lambda row: Age (row),axis=1)
a = sub1.head
print(a)
#new Cancer_Volume variable, categorical 1 through 4
def Cancer_Volume(row):
if row['Cancer Volume'] < 11:
return 1
if row['Cancer Volume'] >11 and row['Cancer Volume'] < 21 :
return 2
if row['Cancer Volume'] >21 and row['Cancer Volume'] <31 :
return 3
if row['Cancer Volume'] >31:
return 4
sub1['Cancer_Volume'] = sub1.apply (lambda row: Cancer_Volume(row),axis=1)
a = sub1.head
print(a)
#univariate bar graph for categorical variables for PSA level
# First hange format from numeric to categorical
sub1["PSA"] = sub1["PSA"].astype('category')
seaborn.countplot(x="PSA", data=sub1)
plt.xlabel('PSA level')
plt.title('PSA level Among Adult men who visited the university medical center in the Prostate cancer Study')
#univariate bar graph for categorical variables for Age groups
# First hange format from numeric to categorical
sub1["AgeF"] = sub1["AgeF"].astype('category')
seaborn.countplot(x="AgeF", data=sub1)
plt.xlabel('AgeF')
plt.title('Age groups Among Adult men who visited the university medical center in the Prostate cancer Study')
#univariate bar graph for categorical variables for Cancer Volume
# First hange format from numeric to categorical
sub1["Cancer_Volume"] = sub1["Cancer_Volume"].astype('category')
seaborn.countplot(x="Cancer_Volume", data=sub1)
plt.xlabel('Cancer_Volume')
plt.title('Cancer Volume Among Adult men who visited the university medical center in the Prostate cancer Study')
# standard deviation and other descriptive statistics for quantitative variables
print ('PSA level')
desc2 = sub1['PSA'].describe()
print (desc2)
c1= sub1.groupby('PSA').size()
print (c1)
print ('mode PSA level')
mode1 = sub1['PSA'].mode()
print (mode1)
c1= sub1.groupby('PSA').size()
print (c1)
p1 = sub1.groupby('PSA').size() * 100 / len(mydata)
print (p1)
# standard deviation and other descriptive statistics for quantitative variables
print ('Age')
desc2 = sub1['AgeF'].describe()
print (desc2)
c2= sub1.groupby('AgeF').size()
print (c2)
print ('mode of Age')
mode1 = sub1['AgeF'].mode()
print (mode1)
p2 = sub1.groupby('AgeF').size() * 100 / len(mydata)
print (p2)
print ('Cancer Volume')
desc2 = sub1['Cancer_Volume'].describe()
print (desc2)
c2= sub1.groupby('Cancer_Volume').size()
print (c2)
print ('Mode of Cancer Volume')
mode1 = sub1['Cancer_Volume'].mode()
print (mode1)
# bivariate bar graph C->Q
seaborn.factorplot(x='AgeF', y='PSA', data=mydata, kind="bar", ci=None)
plt.xlabel('Age')
plt.ylabel('PSA level')
OUTPUT
<bound method NDFrame.head of AgeF PSA Cancer Volume
0 41-50 0.651 0.5599
1 51-60 0.852 0.3716
2 71-80 0.852 0.6005
3 51-60 0.852 0.3012
4 61-70 1.448 2.1170
.. ... ... ...
92 61-70 80.640 16.9455
93 41-50 107.770 45.6042
94 51-60 170.716 18.3568
95 61-70 239.847 17.8143
96 61-70 265.072 32.1367
[97 rows x 3 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume
0 41-50 1 0.5599
1 51-60 1 0.3716
2 71-80 1 0.6005
3 51-60 1 0.3012
4 61-70 1 2.1170
.. ... ... ...
92 61-70 2 16.9455
93 41-50 2 45.6042
94 51-60 2 18.3568
95 61-70 2 17.8143
96 61-70 2 32.1367
[97 rows x 3 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume Age
0 41-50 1 0.5599 1
1 51-60 1 0.3716 2
2 71-80 1 0.6005 4
3 51-60 1 0.3012 2
4 61-70 1 2.1170 3
.. ... ... ... ...
92 61-70 2 16.9455 3
93 41-50 2 45.6042 1
94 51-60 2 18.3568 2
95 61-70 2 17.8143 3
96 61-70 2 32.1367 3
[97 rows x 4 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume Age Cancer_Volume
0 41-50 1 0.5599 1 1
1 51-60 1 0.3716 2 1
2 71-80 1 0.6005 4 1
3 51-60 1 0.3012 2 1
4 61-70 1 2.1170 3 1
.. ... ... ... ... ...
92 61-70 2 16.9455 3 2
93 41-50 2 45.6042 1 4
94 51-60 2 18.3568 2 2
95 61-70 2 17.8143 3 2
96 61-70 2 32.1367 3 4
[97 rows x 5 columns]>
PSA level
count 97
unique 2
top 2
freq 83
Name: PSA, dtype: int64
PSA
1 14
2 83
dtype: int64
mode PSA level
0 2
Name: PSA, dtype: category
Categories (2, int64): [1, 2]
PSA
1 14
2 83
dtype: int64
PSA
1 14.43299
2 85.56701
dtype: float64
Age
count 97
unique 4
top 61-70
freq 59
Name: AgeF, dtype: object
AgeF
41-50 8
51-60 17
61-70 59
71-80 13
dtype: int64
mode of Age
0 61-70
Name: AgeF, dtype: category
Categories (4, object): [41-50, 51-60, 61-70, 71-80]
AgeF
41-50 8.247423
51-60 17.525773
61-70 60.824742
71-80 13.402062
dtype: float64
Cancer Volume
count 97
unique 4
top 1
freq 75
Name: Cancer_Volume, dtype: int64
Cancer_Volume
1 75
2 16
3 4
4 2
dtype: int64
Mode of Cancer Volume
0 1
Name: Cancer_Volume, dtype: category
Categories (4, int64): [1, 2, 3, 4]
The univariate graph of PSA level:
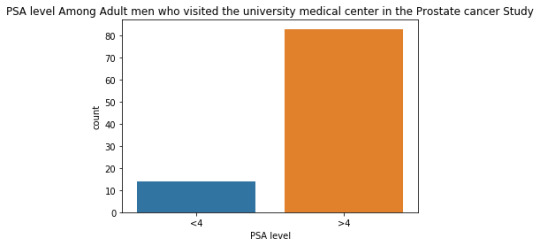
This graph is unimodal, with its highest peak at the category of >4 PSA level . It seems to be skewed to the left as there are higher frequencies in higher category(>4) than the lower category.
The univariate graph of Age groups:
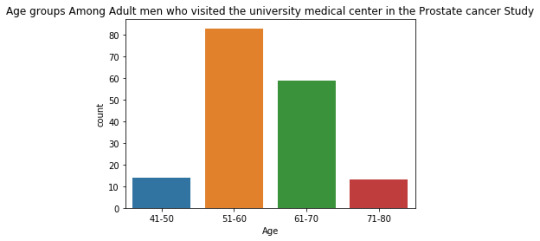
This graph is unimodal, with its highest peak at 51 to 60 age group. It seems to be skewed to the right as there are higher frequencies in the lower age ranges from 51 to 60.
The univariate graph of Cancer Volume:
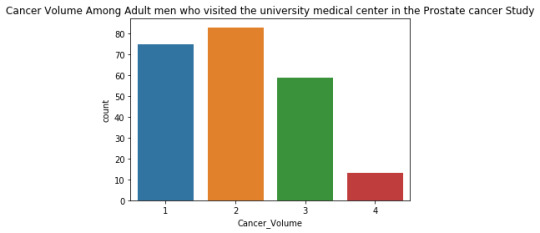
This graph is unimodal, with its highest peak at the category of 2 (11-20) . It seems to be skewed to the right as there are higher frequencies in lower categories than the higher categories.

The graph above plots the Cancer Volume of the adult men to the adult men corresponding Age groups. We can see that the bar chat does not show a clear relationship/trend between the two variables.
0 notes
Link
Bénin / Concours à la CNSS : Le cabinet AGEFIC rejette les allégations de Jean-Baptiste Elias - L'info en temps réel sur Bénin Monde Infos http://beninmondeinfos.com/index.php/benin/21-societe/7443-benin-concours-a-la-cnss-accuse-par-anlc-le-cabinet-agefic-se-defend
0 notes
Text
Week 3
import pandas
import numpy
# any additional libraries would be imported here
mydata = pandas.read_csv('prostate_1.csv', low_memory=False)
# subset variables in new data frame, sub1
sub1=mydata[['AgeF','PSA', 'Cancer Volume']]
a = sub1.head
print(a)
#new PSA variable, categorical 1 through 2
def PSA (row):
if row['PSA'] < 4 :
return 1
if row['PSA'] > 4 :
return 2
sub1['PSA'] = sub1.apply (lambda row: PSA (row),axis=1)
a = sub1.head
print(a)
#new Age variable, categorical 1 through 2
def Age (row):
if row['AgeF']== '41-50':
return 1
if row['AgeF'] == '51-60' :
return 2
if row['AgeF'] == '61-70' :
return 3
if row['AgeF'] == '71-80' :
return 4
sub1['Age'] = sub1.apply (lambda row: Age (row),axis=1)
a = sub1.head
print(a)
#new Cancer_Volume variable, categorical 1 through 4
def Cancer_Volume(row):
if row['Cancer Volume'] < 11:
return 1
if row['Cancer Volume'] >11 and row['Cancer Volume'] < 21 :
return 2
if row['Cancer Volume'] >21 and row['Cancer Volume'] <31 :
return 3
if row['Cancer Volume'] >31:
return 4
sub1['Cancer_Volume'] = sub1.apply (lambda row: Cancer_Volume(row),axis=1)
a = sub1.head
print(a)
#frequency distributions for primary and secondary ethinciity variables
print( 'counts for PSA level')
c10 = sub1['PSA'].value_counts(sort=False)
print(c10)
print( 'percentages for PSA level')
p10 = sub1['PSA'].value_counts(sort=False, normalize=True)
print (p10)
print('counts for Age')
c11 = sub1['Age'].value_counts(sort=False)
print(c11)
print( 'percentages for Age')
p11= sub1['Age'].value_counts(sort=False, normalize=True)
print (p11)
print( 'counts for Cancer Volume')
c12 = sub1['Cancer_Volume'].value_counts(sort=False)
print(c12)
print( 'percentages for Cancer Volume')
p12 = sub1['Cancer_Volume'].value_counts(sort=False, normalize=True)
print (p12)
Output
runfile('C:/Users/NAOMI/Downloads/Documents/Cousera/Data Visualization/week 3/Assignment 3 new.py', wdir='C:/Users/NAOMI/Downloads/Documents/Cousera/Data Visualization/week 3')
<bound method NDFrame.head of AgeF PSA Cancer Volume
0 41-50 0.651 0.5599
1 51-60 0.852 0.3716
2 71-80 0.852 0.6005
3 51-60 0.852 0.3012
4 61-70 1.448 2.1170
.. ... ... ...
92 61-70 80.640 16.9455
93 41-50 107.770 45.6042
94 51-60 170.716 18.3568
95 61-70 239.847 17.8143
96 61-70 265.072 32.1367
[97 rows x 3 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume
0 41-50 1 0.5599
1 51-60 1 0.3716
2 71-80 1 0.6005
3 51-60 1 0.3012
4 61-70 1 2.1170
.. ... ... ...
92 61-70 2 16.9455
93 41-50 2 45.6042
94 51-60 2 18.3568
95 61-70 2 17.8143
96 61-70 2 32.1367
[97 rows x 3 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume Age
0 41-50 1 0.5599 1
1 51-60 1 0.3716 2
2 71-80 1 0.6005 4
3 51-60 1 0.3012 2
4 61-70 1 2.1170 3
.. ... ... ... ...
92 61-70 2 16.9455 3
93 41-50 2 45.6042 1
94 51-60 2 18.3568 2
95 61-70 2 17.8143 3
96 61-70 2 32.1367 3
[97 rows x 4 columns]>
<bound method NDFrame.head of AgeF PSA Cancer Volume Age Cancer_Volume
0 41-50 1 0.5599 1 1
1 51-60 1 0.3716 2 1
2 71-80 1 0.6005 4 1
3 51-60 1 0.3012 2 1
4 61-70 1 2.1170 3 1
.. ... ... ... ... ...
92 61-70 2 16.9455 3 2
93 41-50 2 45.6042 1 4
94 51-60 2 18.3568 2 2
95 61-70 2 17.8143 3 2
96 61-70 2 32.1367 3 4
[97 rows x 5 columns]>
counts for PSA level
1 14
2 83
Name: PSA, dtype: int64
percentages for PSA level
1 0.14433
2 0.85567
Name: PSA, dtype: float64
counts for Age
1 8
2 17
3 59
4 13
Name: Age, dtype: int64
percentages for Age
1 0.082474
2 0.175258
3 0.608247
4 0.134021
Name: Age, dtype: float64
counts for Cancer Volume
1 75
2 16
3 4
4 2
Name: Cancer_Volume, dtype: int64
percentages for Cancer Volume
1 0.773196
2 0.164948
3 0.041237
4 0.020619
Name: Cancer_Volume, dtype: float64
I created new data with three variables: AgeF, PSA and Cancer Volume. The were no missing data set in my data. For Age, the most commonly endorsed is 3 (60.8%) , meaning more than half of the men who went for the checkup are from the age 61-70 years. For PSA, 2 (85.57%) has the highest percentage, meaning the PSA which is greater than 4 has the highest frequency of 83. For Cancer Volume, 1 ( 77.32% ) has the highest percentage among the others which means the Cancer Volume less than 10 has the highest frequency of 75 with 77.32%.
0 notes