#DNABarcoding
Explore tagged Tumblr posts
Text
Universal primer untuk identifikasi Bakteri: Barcoding dan Metabarcoding
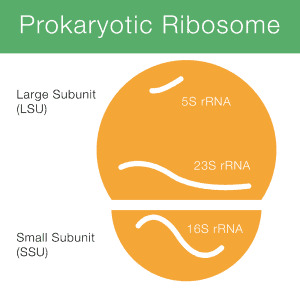
16S rRNA adalah singkatan dari 16S ribosomal ribonucleic acid (rRNA), di mana S (Svedberg) merupakan satuan pengukuran (laju sedimentasi). rRNA ini merupakan komponen penting dari subunit kecil (SSU) ribosom prokariot, serta terdapat dalam mitokondria dan kloroplas. Gambar 1 menunjukkan bagaimana 16S rRNA (disingkat sebagai 16S) berperan dalam ribosom prokariot. Pengurutan gen 16S rRNA merupakan…
View On WordPress
0 notes
Text
The Eye of Sauron in the Amazon
ITA version ESP version
It is not always easy to agree on a name for new species, but researchers at the Natural History Museum in London had no doubts when they encountered new species of vegetarian piranhas.
But let’s take a step back. In the past, defining an animal as belonging to a new species often relied on morphological characteristics. This is the case with the genus Myloplus, particularly the species Myloplus schomburgkii. Under this name, fish were grouped together that could be easily recognised by their vertical black bar in the centre of the body and their diet mainly consisting of plants and some insects, despite their close relationship to piranhas. Through a new and careful morphological analysis of multiple specimens combined with DNA barcoding, researchers realised that within the species Myloplus schomburgkii, there were actually two other species, all with a dark vertical bar on the side, but genetically distinct!
The two new species were named M. aylan and M. sauron. The specific name M. aylan honours the late Aylan Moraes Andrade, the son of Carine Moraes and Marcelo Andrade, one of the authors, who passed away prematurely. The specific name M. sauron alludes to the Eye of Sauron from J. R. R. Tolkien’s “The Lord of the Rings” as its body is marked with a vertical black bar tapering towards both ends, resembling the famous vertical-pupilled eye from the novel. Additionally, some specimens also have orange spots on their bodies, just like the Eye!
How did the researchers realise they were dealing with multiple species? They found significant molecular distances between the three lineages, well beyond the minimum threshold for fish species DNA barcoding (2%-3%). Myloplus schomburgkii differs from M. aylan and M. sauron by 7.9% and 9.7% respectively. The distance between M. aylan and M. sauron was even greater, at 11%.
The differences between the three species are not limited to genetics. To a keen eye, the same dark vertical bar can be easily used to distinguish the three species. Myloplus schomburgkii has a uniformly wide vertical dark bar compared to the two new species. In M. sauron, the vertical black bar tapers towards the ends forming tapered distal points, while in M. aylan, the vertical dark bar has a broader region near the lateral line and the ends do not taper.
The naming of the new species M. aylan and M. sauron reflects the deep connection between science and society, where every name tells a story. This discovery, made possible through a combination of morphological and genetic analyses, reminds us that our understanding of the natural world is continuously evolving. Nature always surprises us, and the identification of these new species highlights how vast and still partly unknown the animal kingdom is. The researchers invite us to maintain a watchful and curious eye, just like the Eye of Sauron, to continue exploring and discovering the wonders of nature.
Source

#EyeOfSauron#Amazon#NewSpecies#VegetarianPiranhas#NaturalHistoryMuseum#London#Myloplus#Biodiversity#ScientificDiscovery#MorphologicalAnalysis#DNABarcoding#Genetics#Conservation#ScientificResearch#Nature#Exploration#Myloplus Schomburgkii#Myloplus Aylan#Myloplus Sauron#StoriesOfNature#LordOfTheRings#JRRTolkien#Animals#Evolution#ScienceAndSociety#WondersOfNature
0 notes
Text
Abstract The Crucifers family (Brassicaceae) includes a large number of economically important crops, particularly Brassica rapa, which is a widely used model plant for molecular genetic studies of oilseeds. B. rapa is a highly polymorphic species that includes many of genetically distinct subspecies. Considering this fact, the intraspecific hybridization of B. rapa subspecies is considered a promising breeding approach aimed at increasing the genetic diversity of the crop. Previously, the authors have shown that one of such hybrids, B. rapa subsp. oleifera f. biennis × (subsp. rapifera × pekinensis), could be a valuable oil feedstock due to its increased productivity. However, obtaining hybrids and their subsequent breeding would require the involvement of various molecular marker systems. So far, the method of estimating the length polymorphism of the first (TBP) and second (cTBP) introns of β-tubulin has demonstrated its high accuracy and reliability in the identification (DNA-barcoding) of flowering plant taxonomic units at different levels. In the present study, the productivity of such hybrid oil tyfon (B. rapa subsp. oleifera f. biennis × (subsp. rapifera × pekinensis)) was evaluated and DNA-barcoding of different hybrid tyfon lines (B. rapa subsp. oleifera f. biennis × (subsp. rapifera × pekinensis)) and its parental B. rapa subspecies using the β-tubulin intron length polymorphism assessment approach was carried out. Based on the data of the molecular genetic analysis, which included the assessment of length polymorphism of the first and second introns of β-tubulin genes, we were able to confirm the origin of the oil tyfon hybrid (B. rapa subsp. oleifera f. biennis × (subsp. rapifera × pekinensis)) hybrid from Dutch leaf tyfon (B. rapa subsp. rapifera × pekinensis) and winter turnip (B. rapa subsp. oleifera) with high confidence. Along with that, it was possible to differentiate var. glabra and var. laxa accession of napa cabbage (B. rapa subsp. pekinensis) for the first time using combined TBP and cTBP analyses. A variation in the number of amplified regions of β-tubulin introns was noted in different genotypes; however, these differences did not appear to be a specific feature of a particular subspecies/hybrid. This suggests that B. rapa hybrids most likely do not differ in ploidy compared to their parental genotypes. In addition, it was shown that the mentioned oil tyfon hybrid lines of Ukrainian breeding show a significant level of morphological variation despite their common breeding pedigree.
0 notes
Link
Face IT DNA Technology has very good and strong relationships with hundreds of hospitals and clinics all over the country. It means you are able to schedule a personal visit with a skilled and professional healthcare provider to have DNA paternity testing completed just about anywhere nearby your location.
0 notes
Text
Conference: South Africa, 7th International Barcode of
7th International Barcode of Life Conference, 20 – 24 November 2017 at Kruger National Park The African Centre for DNA Barcoding (ACDB), the University of Johannesburg (UJ), International Barcode of Life Project (iBOL), and the Department of Environmental Affairs (DEA) are proud to announce and welcome delegates to our hosting of the 7th International Barcode of Life Conference, 20 – 24 November 2017. This is the first time that this event will be held on the African continent. The venue for the hosting of this prestigious event will be the Nombolo Mdhluli Conference Centre, Skukuza. The major theme of the conference is exploring mega-diverse biotas with DNA barcodes. A series of presentations and workshops will focus on the use of DNA to understand diversity patterns and ecological processes in species-rich and complicated ecosystems. The conference also provides a general forum for presentations, posters, and discussion on the wider field of DNA barcoding. Delegates are encouraged to register as soon as possible as space is limited. Abstracts should be submitted at http://bit.ly/2o0KXYJ before or on 14th April 2017. More information at: Website: http://bit.ly/2nfl6z4 Facebook: @DNABarcodes2017 Twitter: @DNABarcodes via Gmail
0 notes
Quote
RT @DNABARCODE: Help us out? #WATCH https://t.co/GsIvyuEoU8 & give it a thumbs up $10K prize from #MedTechInnovator! #TY!… https://t.co/SyV4UMUkf4
http://twitter.com/Cassie
0 notes
Text
RT https://t.co/tecI1akpSm RT @DNABARCODE: Triple Negative #Breast #OneRNA provides treatment options whe… https://t.co/5RYWa4GXG7
RT https://t.co/tecI1akpSm RT @DNABARCODE: Triple Negative #Breast #Cancer #OneRNA provides treatment options whe… https://t.co/5RYWa4GXG7
— Mesothelioma Explain (@mesotheliomaex) July 25, 2017
Source: @mesotheliomaex July 25, 2017 at 11:55AM More info What Is Mesothelioma Cancer
#What Is Mesothelioma Cancer#Mesothelioma Cell Types#Mesothelioma Surgery Options#Mesothelioma Misdia
0 notes
Text
Tweeted
The latest The Business Trends Daily! https://t.co/HUlrsKYj5z Thanks to @joshspilker @pitchpivot @DNABARCODE #startups #entrepreneur
— Cory C. Claytor (@CoryCClaytor) April 26, 2017
0 notes
Photo

How diverse is our school’s biological community?
This year we participated in the School Malaise Trap Program hosted by Biodiversity Institute of ON. I first learned about this program at STAO in 2014 and was so excited to be a part of it this year. It fit in nicely with our theme of citizen science in grade 9. The program allows students to become part of the broader scientific community by conducting authentic inquiry. The program sent us a malaise trap in order to collect samples of insects in our yard. We then sent the samples back to the Biodiversity Institute where they were DNA barcoded. Our results indicated that despite our urban school setting, the insect community was more diverse than expected! Our trap collected a total of 101 specimens in week 1 and 118 specimens in week 2. Within the sample, there were 72 species, which ranked our sample 36 out of 64 for species diversity. The species collected came from several different groups and 13 species were only collected in our trap. We also collected one species of hymenoptera that was NEW to the Barcode Of Life Database! Amazing! I will definitely sign up for this program again - as soon as our renovations are done and we have our green space back!
0 notes
Text
L’Occhio di Sauron dell’Amazzonia
ENG version ESP version Non sempre è facile mettersi d’accordo in gruppo quando si cerca il nome per nuove specie, ma i ricercatori del Museo di Storia Naturale di Londra non hanno avuto dubbi quando si sono trovati davanti a nuove specie di piranha vegetariani.
Ma facciamo un passo indietro. Per definire un animale come appartenente a una specie nuova, in passato spesso ci si basava sulle caratteristiche morfologiche. Questo è il caso del genere Myloplus, e in particolare della specie Myloplus schomburgkii. Sotto questo nome, infatti, venivano racchiusi pesci facilmente riconoscibili per avere una barra nera verticale al centro del corpo e per la particolare dieta basata soprattutto su piante, e qualche insetto, nonostante la loro stretta parentela coi piranha. I ricercatori, attraverso una nuova e attenta analisi morfologica di più esemplari unita alla codifica a barre del DNA, si sono resi conto che all’interno della specie Myloplus schomburgkii si nascondevano in realtà altre due specie, tutte con una barra verticale scura sul fianco, ma geneticamente diverse!
Alle due nuove specie sono stati dati rispettivamente i nomi di M. aylan e M. sauron. Il nome specifico M. aylan onora il defunto Aylan Moraes Andrade, figlio di Carine Moraes e Marcelo Andrade, uno degli autori, scomparso prematuramente. Il nome specifico M. sauron, invece, allude all'Occhio di Sauron, dal “Signore degli Anelli” di J. R. R. Tolkien in quanto il suo corpo è segnato con una barra verticale nera che si restringe verso entrambe le estremità, che ricorda il famoso occhio dalla pupilla verticale del romanzo. Inoltre, alcuni esemplari presentano anche delle macchie arancioni sul corpo, proprio come l’Occhio!
Come si sono resi conto i ricercatori di trovarsi davanti a più specie? Ebbene, loro hanno riscontrato distanze molecolari tra i tre lignaggi piuttosto elevate che andavano ben oltre la soglia minima per il DNA Barcoding delle specie ittiche (dal 2% al 3%). Myloplus schomburgkii differisce da M. aylan e M. sauron rispettivamente del 7,9% e del 9,7%. Tra M. aylan e M. sauron la distanza era ancora maggiore, 11%. Le differenze tra le tre specie non si fermano alla parte genetica. A un occhio più acuto, la stessa barra scura verticale può essere facilmente utilizzata per distinguere le tre specie. Myloplus schomburgkii presenta una barra verticale scura di larghezza uniforme rispetto alle due nuove specie. In M. sauron la barra nera verticale si restringe verso le estremità formando punte distali affusolate, mentre in M. aylan la barra scura verticale ha una regione più ampia vicino alla linea laterale e le estremità non si assottigliano.
La denominazione delle nuove specie M. aylan e M. sauron riflette la profondità del legame tra scienza e società, dove ogni nome racconta una storia. Questa scoperta, resa possibile grazie alla combinazione di analisi morfologiche e genetiche, ci ricorda che la nostra comprensione del mondo naturale è in continua evoluzione. La natura ci sorprende sempre, e l'identificazione di queste nuove specie sottolinea quanto sia vasto e ancora in parte sconosciuto il regno animale. I ricercatori ci invitano a mantenere uno sguardo attento e curioso, proprio come l'Occhio di Sauron, per continuare a esplorare e scoprire le meraviglie della natura.
Fonte articolo e foto

#Amazzonia#NuoveSpecie#PiranhaVegetariani#MuseoDiStoriaNaturale#Londra#Myloplus#Biodiversità#ScopertaScientifica#AnalisiMorfologica#DNABarcoding#Genetica#Conservazione#RicercaScientifica#Natura#Esplorazione#Myloplus Schomburgkii#Myloplus Aylan#Myloplus Sauron#StorieDellaNatura#OcchioDiSauron#SignoreDegliAnelli#JRRRTolkien#Animali#SpecieNuove#Evoluzione#ScienzaESocietà#MeraviglieDellaNatura#occhio di sauron
0 notes
Link
With the Pro Package it goes into even further details. As you may have learned back in science or biology class, generally a cleft chin is something that’s inherited as well as dimples, widow’s peak, freckles and other hereditary characteristics. facednatest.com allows you to check blood types. Read more here … https://bit.ly/2Wf2KLB.
0 notes
Link
Founders and inventors of automated face recognition include Helen Chan Wolf, Woody Bledsoe and Charles Bisson and Lawrence Reese
0 notes
Text
RT https://t.co/BGJweaWIRU RT @DNABARCODE: Support me #Paddleboarding to raise funds for The #Breast Rese… https://t.co/yQWhMXdiS5
RT https://t.co/BGJweaWIRU RT @DNABARCODE: Support me #Paddleboarding to raise funds for The #Breast #Cancer Rese… https://t.co/yQWhMXdiS5
— Mesothelioma Explain (@mesotheliomaex) July 25, 2017
Source: @mesotheliomaex July 25, 2017 at 11:55AM More info What Is Mesothelioma Cancer
#What Is Mesothelioma Cancer#Mesothelioma Cell Types#Mesothelioma Surgery Options#Mesothelioma Misdia
0 notes