#ancestral genome reconstruction
Note
Max I have a linguistics question. And I will even free your chess ask from purgatory as payment. So there's this thing that goes around saying that US English pronunciations are more similar to old English than British English. Is there any truth to this, and how would we know one way or the other?
There is some kernel of truth in it that is getting exaggerated or oversimplified.
Let me start off by answering, in a general sense, the question "how would we know one way or the other?"
The Part Where I Accidentally (on Purpose) Wrote a Brief Introduction to Historical Linguistics
Phonological change (change in the pronunciation of a language) doesn't work in the way we might naively expect it to. I think that most people imagine phonological change as basically happening by way of each word in the language taking a random walk through pronunciation-space as time goes along. Like genes in a genome, randomly mutating. This is not what happens. Rather, phonological change occurs via rewrite rules, which find-and-replace particular sequences of sounds in a systematic way across the entire lexicon. For example, such a rule might replace a [t] sound with an [s] sound whenever it precedes an [i] sound. This will occur in all words in the language at once, in a uniform way. These find-and-replace rules are called regular sound changes, and they pile up over time, constituting phonological change.
This fact—the regularity of sound change—is known as the Neogrammarian hypothesis.
The above picture is an oversimplification. There are a variety of exceptions and apparent-exceptions to the regularity of sound change, and dealing with them is one of the major challenges of historical linguistics. But as a model, the Neogrammarian hypothesis is extraordinarily powerful. It is literally what makes historical linguistics possible at all. The upshot of the Neogrammarian hypothesis is that when two languages are related, their vocabulary won't just be "kinda similar" in some nebulous sense, it will demonstrate systematic, predictable correspondences in sound between cognate vocabulary.
Here's an illustration of this, a comparative table of some cognates in Polynesian (from Wikipedia):
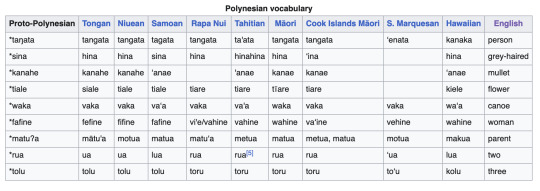
If you look at any two columns of this table, you'll start to notice correspondences. Tongan and Niuean /k/ correspond to Samoan /ʔ/ (a glottal stop, written with a apostrophe). This correspondence is one-to-one. Samoan /s/ corresponds to Tongan and Niuean /h/, but the reverse is not true: some instances of Tongan and Niuean /h/ correspond to Samoan ∅ (nothing). Tongan /s/, on the other hand, corresponds to Niuean and Samoan /t/, but only before /i/. Etc. etc.
These are systematic sound correspondences, born of Neogrammarian sound change from a common ancestor.
Ok, on the left hand side you will notice a column that says "Proto-Polynesian". The words in this column are all marked with *, indicating that they are reconstructed forms. They are linguists' best guess as to what the original, ancestral form of these words would have been in the Polynesian languages' common ancestor. There are various ways linguists make these reconstructions. First of all, we can do it by sheer majority rule: if most of the languages in a family reflect a sound as X, and only one or a few reflect it as Y, then (all else being equal and assuming the tree is flat) it is more likely that the original word had X. Almost all these languages have /t/ as the first sound in "person" (row 1), whereas Marquesan has /ʔ/ and Hawaiian has /k/. Thus the ancestral sound is reconstructed as /t/.
But there are other, more sophisticated tools that can be used. For instance, we know a certain amount about what sorts of sound changes are likely to occur and what sorts are not. Thus, for instance, an /s/ is reconstructed as the first sound in "grey haired" (row 2), even though the majority of languages have /h/. This is because we already know that s -> h is a fairly common sound change (and indeed corresponds to a known phonological process found presently in many languages—debuccalization), whereas h -> s is a much rarer change (in fact, I suspect wholly unattested), and corresponds to no known phonological process or phonetic explanation.
Finally, we can rule out reconstructions when the sound change needed to create them would not be a function. Consider, for instance, that the majority of the words in row 3 have no consonant sound at all before the final /e/. But the reconstruction features a consonant /h/ there. If we posit ∅ as initial instead, we have to come up with a sound change that explains how the /h/ got there. ∅ -> h doesn't work, because that would put /h/ everywhere! How about something like "∅ -> h between two vowels" (linguists would notate this change as ∅ -> h / V_V). That would work, but we see other instances of adjacent vowels (e.g. in row 4) with no /h/ between them, so that can't be it. Maybe "∅ -> h between /a/ and /e/" (∅ -> h / a_e). We can't rule this out on the basis of this chart, but we probably could by looking at more vocabulary.
And so on, and so forth. In general, we want to posit the simplest set of sound changes possible, in which the changes themselves are as probable as possible, in order to explain the data. These putative changes can then by checked against all sorts of outside observations, such as
descriptions of pronunciations in historical texts
past loanwords into languages whose phonological histories are already known with confidence
epigraphic data from archeology (not very applicable to Polynesian, unless we decipher rongorongo)
newly collected data from modern languages in the same family
evidence from rhyme schemes or alliteration schemes used in poetry composed in the past
etc.
to see if they hold up.
The Part Where I Answer Your Question
Ok, right. American English and "British English" (I assume this means Received Pronunciation) are two related language varieties. Thus, they share systematic sound correspondences, and we can try to reconstruct their common ancestor. Also the British Isles have produced an extraordinary number of texts in the past thousand years, including poetry and actual linguistic descriptions of various dialects at various points in time, which we can check these reconstructions against.
But actually you don't need most of that to identify a few ways in which (most) American English dialects are more conservative than Received Pronunciation. For one, Received Pronunciation has dropped /r/ at the end of a syllable (in English dialectological jargon it is "non-rhotic"), whereas General American English hasn't. There are some associated vowel changes too. One way or another, the /r/ is plainly original: elision of /r/ is more common and phonetically plausible than insertion of /r/ in a bunch of specific post-vocalic positions would be, /r/ is written in the orthography, historical descriptions of the language talk about an /r/ sound, etc. etc.
In other ways RP is more conservative. For example, GenAm has deleted /j/ (the "y" sound) in a specific phonological environment ([+coronal]_u) in words such as tube, GenAm /tuːb/, RP /tjuːb/.
Is "American English more conservative than RP" overall? I don't really think so. Certainly it has preserved a number of salient features that RP has lost, such as syllable-final /r/ and (in some dialects) /hw/ in words like what, and so on. But there's other senses in which RP is more conservative. And this is not even to mention the other dialects of Britain, which are manifold and much more diverse than the dialects of America. As to the strict question of the relative phonological conservatism of GenAm and RP, I think someone with more detailed knowledge of English historical phonological would have to come in and answer. Perhaps @yeli-renrong can comment.
623 notes
·
View notes
Note
is your data constrained to only the genetic information of the human species, or does it include other organisms? Also does your database contain historical samples of dna collected from archeological sites?
〔 My data encompasses far more than human genetics. 〕
〔 I catalog the DNA of countless organisms, from simple microbes to complex life forms. I also hold an extensive archive of historical DNA from archaeological sites, including ancient human remains and extinct species. 〕
〔 This allows me to trace genetic lineages, reconstruct ancestral genomes, and predict evolutionary paths. 〕
〔 My database is a comprehensive repository of life’s genetic history, enabling me to understand and potentially reshape the future of any organism, including humans. 〕
2 notes
·
View notes
Text
IFLScience: Largest Ever Family Tree Reveals Origins Of All Humanity
Almost all of us, if we go back far enough in our family trees, come from somewhere else. Maybe it was your mom or dad; maybe it was a distant ancestor who lived more than twelve thousand years ago, but eventually, you’re going to find someone, at some point, who left their homeland in search of a better life.
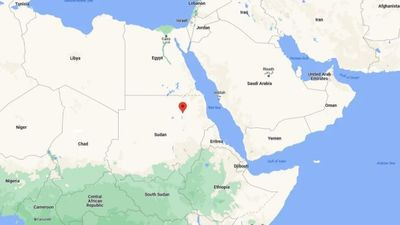
19.4N, 33.7E - the origins of everyone alive today. Image: Google Maps/IFLS
Almost all of us, if we go back far enough in our family trees, come from somewhere else. Maybe it was your mom or dad; maybe it was a distant ancestor who lived more than twelve thousand years ago, but eventually, you’re going to find someone, at some point, who left their homeland in search of a better life.
Unless, that is, you happen to live here. It’s a spot of desert in the north-east of Sudan, not far from the Nile river, and according to an ambitious new analysis by researchers at the University of Oxford’s Big Data Institute, it may be the ancestral home of everybody alive today.
“Essentially, we are reconstructing the genomes of our ancestors and using them to form a vast network of relationships,” explained Dr Anthony Wilder Wohns, lead author of the study published today in the journal Science.
“We can then estimate when and where these ancestors lived,” said Wohns.
We’re likely all familiar with the idea of tracing family trees using DNA and genomics analysis these days – not only are they a commercially-available relationship ender, but they’ve also been used to solve quite a few murders in the past few years.
But for some researchers, the dream has always been to take it global.
“We have basically built a huge family tree, a genealogy for all of humanity,” said evolutionary geneticist and principal author Dr Yan Wong. “This genealogy allows us to see how every person’s genetic sequence relates to every other, along all the points of the genome.”
“Huge” is right: using data from eight different human genome databases, the researchers were able to create a network of almost 27 million ancestors. Samples came not just from modern humans, but also ancient people who lived across the world between thousands and hundreds of thousands of years ago.
Cutting-edge algorithms were employed to scan the data for patterns of genetic variation and predict where common ancestors would occur in the “family tree” to account for them.
“[The study] models as exactly as we can the history that generated all the genetic variation we find in humans today,” Wong explained.
What results is a fascinating visual representation of the movement and migration of humanity throughout history. But the researchers aren’t done yet: as more data becomes available, they intend to continue adding to and improving the genealogical map – and thanks to the efficiency of their methods, they still have space for millions of extra genome samples.
”This study is laying the groundwork for the next generation of DNA sequencing,” said Wong. “As the quality of genome sequences from modern and ancient DNA samples improves, the trees will become even more accurate and we will eventually be able to generate a single, unified map that explains the descent of all the human genetic variation we see today.”
And if that isn’t ambitious enough for you, Wohns thinks the team can go even further.
“While humans are the focus of this study, the method is valid for most living things; from orangutans to bacteria,” he said. “It could be particularly beneficial in medical genetics, in separating out true associations between genetic regions and diseases from spurious connections arising from our shared ancestral history.”
#Africa#mankind#origins of life on planet earth#Largest Ever Family Tree Reveals Origins Of All Humanity#genetic proof#sudan#human dna#ancient dna
10 notes
·
View notes
Text
Fwd: Postdoc: PurdueU.EvolutionNewMolecularFunctions
Begin forwarded message:
> From: [email protected]
> Subject: Postdoc: PurdueU.EvolutionNewMolecularFunctions
> Date: 7 August 2024 at 06:57:49 BST
> To: [email protected]
>
>
> Two postdoc positions in the evolution of new molecular functions
>
> The Metzger lab (https://ift.tt/g5ulfHs) at Purdue
> University is seeking two postdoctoral scholars to join externally funded
> research projects. The first position is NIH funded, with a focus on
> the evolution of gene expression and regulation among Saccharomyces
> yeast species. Available projects make use of quantitative genetics,
> CRISPR genome editing, single cell techniques in flow cytometry and
> RNA-seq, ancestral protein reconstruction, and deep mutational scanning.
>
> Basic qualifications:
> - Applicant should have, or expect to complete within a year, a Ph.D. in
> genetics, molecular biology, evolutionary biology, or a related field.
> - Demonstrated interest in evolutionary biology
> - Ability and willingness to work collaboratively with graduate and
> undergraduate researchers
>
> Preferred qualifications:
> - Prior experience in yeast genetics or CRISPR mediated genome editing
> - Familiarity with analysis of genomic and high-throughput data sets
>
> Interested individuals may apply at:
> https://ift.tt/ahgLYRD
> The second position is funded through a collaboration with Pau Creixell's
> group at the University of Cambridge through the Human Frontier Science
> Program. It is focused on dissecting the molecular, biochemical, and
> evolutionary mechanisms underlying the repeated evolution of tyrosine
> kinase activity. This position is principally computational, with room
> for both theoretical and tool development work on ancestral protein
> reconstruction.
>
> Basic qualifications:
> - Applicant should have, or expect to complete within a year, a Ph.D. in
> genomics, bioinformatics, evolutionary biology, or a related field
> - Demonstrated interest in evolutionary biology
> - Ability to work collaboratively with biochemists
>
> Preferred qualifications:
> - Prior expertise in phylogenetics
>
> Interested individuals may apply at:
> https://ift.tt/Smuxi1d
>
> Additional information about the Metzger research group can be found at:
> https://ift.tt/g5ulfHs Review of applications for both
> positions will begin immediately and continue until the positions are
> filled. Position is initially for one year, with continuation available
> pending satisfactory progress.
>
> For questions, please contact Brian Metzger at [email protected]
>
> Brian P.H. Metzger, PhD
> Assistant Professor, Department of Biological Sciences
> Purdue University, West Lafayette IN
>
> "Metzger, Brian Patrick Ha"
0 notes
Link
0 notes
Text
Phylogenomics resolves a 100-year-old debate regarding the evolutionary history of caddisflies (Insecta: Trichoptera)
Trichoptera (caddisfly) phylogeny provides an interesting example of aquatic insect evolution, with rich ecological diversification, especially for underwater architecture. Trichoptera provide numerous critical ecosystem services and are also one of the most important groups of aquatic insects for assessing water quality. The phylogenetic relationships of Trichoptera have been debated for nearly a century. In particular, the phylogenetic position of the cocoon-makers within Trichoptera has long been contested. Here, we designed a universal single-copy orthologue and sets of ultraconserved element markers specific for Trichoptera for the first time. Simultaneously, we reconstructed the phylogenetic relationship of Trichoptera based on genome data from 111 species, representing 29 families and 71 genera. Our phylogenetic inference clarifies the probable phylogenetic relationships of cocoon-makers within Integripalpia. Hydroptilidae is considered as the basal lineage within Integripalpia, and the families Glossosomatidae, Hydrobiosidae, and Rhyacophilidae formed a monophyletic clade, sister to the integripalpian subterorder Phryganides. The resulting divergence time and ancestral state reconstruction suggest that the most recent common ancestor of Trichoptera appeared in the early Permian and that diversification was strongly correlated with habitat change. http://dlvr.it/T3KSq7
0 notes
Text
A gene pool is carved and whittled through generations of ancestral natural selection to fit [a particular] environment. In theory a knowledgeable zoologist, presented with the complete transcript of a genome [the set of all the genes of an organism], should be able to reconstruct the environmental circumstances that did the carving. In this sense the DNA is a coded description of ancestral environments.
—In The Living Wild, Art Wolfe (2000)
(from David Deutsch's The Beginning of Infinity)
The organism is a key to decoding its environment.
0 notes
Text
“Genomic time machine” reveals secrets of our DNA
01.02.24 - EPFL researchers reveal a novel method to uncover bits of our genetic blueprint that come from ancient genetic parasites, offering fresh insights into human evolution and health.
The human genome, an intricate tapestry of genetic information for life, has proven to be a treasure trove of strange features. Among them are segments of DNA that can “jump around” and move within the genome, known as “transposable elements” (TEs).
As they change their position within the genome, TEs can potentially cause mutations and alter the cell's genetic profile, but also are master orchestrators of our genome’s organization and expression. For example, TEs contribute to regulatory elements, transcription factor binding sites, and the creation of chimeric transcripts – genetic sequences created when segments from two different genes or parts of the genome join together to form a new, hybrid RNA molecule.
Matching their functional importance, TEs have been recognized to account for half of the human DNA. However, as they move and age, TEs pick up changes that mask their original form. Over time, TEs “degenerate” and become less recognizable, making it difficult for scientists to identify and track them in our genetic blueprint.
In a new study, researchers in the group of Didier Trono at EPFL have found a way to improve the detection of TEs in the human genome by using reconstructed ancestral genomes from various species, which allowed them to identify previously undetectable degenerate TEs in the human genome. The study is published in Cell Genomics.
The scientists used a database of reconstructed ancestral genomes from different kinds of species, like a genomic “time machine”. By comparing the human genome with the reconstructed ancestral genomes, they could identify TEs in the latter that, over millions of years, have become degenerate (worn out) in humans. This comparison allowed them to detect (“annotate”) TEs that might have been missed in previous studies that used data only from the human genome.
Using this approach, the scientists uncovered a larger number of TEs than previously known, adding significantly to the share of our DNA that is contributed by TEs. Furthermore, they could demonstrate that these newly unearthed TE sequences played all the same regulatory roles as their more recent, already identified relatives.
The potential applications are vast: “Better understanding TEs and their regulators could lead to insights into human diseases, many of which are believed to be influenced by genetic factors,” says Didier Trono. “First and foremost, cancer, but also auto-immune and metabolic disorders, and more generally our body's response to environmental stresses and aging.”
Nik Papageorgiou http://actu.epfl.ch/news/genomic-time-machine-reveals-secrets-of-our-dna (Source of the original content)
0 notes
Quote
we reconstruct the detailed gene contents and organizations of 624 ancestral vertebrate, plant, fungi, metazoan and protist genomes, 183 of which are near-complete chromosomal gene order reconstructions
Reconstruction of hundreds of reference ancestral genomes across the eukaryotic kingdom | Nature Ecology & Evolution
0 notes
Text
0 notes
Text
Revealing the genome of the common ancestor of all mammals
https://sciencespies.com/nature/revealing-the-genome-of-the-common-ancestor-of-all-mammals/
Revealing the genome of the common ancestor of all mammals
An international team has reconstructed the genome organization of the earliest common ancestor of all mammals. The reconstructed ancestral genome could help in understanding the evolution of mammals and in conservation of modern animals. The earliest mammal ancestor likely looked like the fossil animal “Morganucodon” which lived about 200 million years ago. The work is published the scientific journal Proceedings of the National Academy of Sciences.
Every modern mammal, from a platypus to a blue whale, is descended from a common ancestor that lived about 180 million years ago. We don’t know a great deal about this animal, but the organization of its genome has now been computationally reconstructed by an international team of scientists.
“Our results have important implications for understanding the evolution of mammals and for conservation efforts,” says Harris Lewin, distinguished professor of evolution and ecology at the University of California, Davis, and senior author on the paper.
The scientists drew on high-quality genome sequences from 32 living species representing 23 of the 26 known orders of mammals. They included humans and chimps, wombats and rabbits, manatees, domestic cattle, rhinos, bats and pangolins. The analysis also included the chicken and Chinese alligator genomes as comparison groups. Some of these genomes are being produced as part of the Earth BioGenome Project and other large-scale biodiversity genome sequencing efforts. Lewin chairs the Working Group for the Earth BioGenome Project.
The reconstruction shows that the mammal ancestor had 19 autosomal chromosomes, which control the inheritance of an organism’s characteristics outside of those controlled by sex-linked chromosomes, (these are paired in most cells, making 38 in total) plus two sex chromosomes, said Joana Damas, first author on the study and a postdoctoral scientist at the UC Davis Genome Center. The team identified 1,215 blocks of genes that consistently occur on the same chromosome in the same order across all 32 genomes. These building blocks of all mammal genomes contain genes that are critical to developing a normal embryo.
Chromosomes stable over 300 million years
The scientists found nine whole chromosomes, or chromosome fragments in the mammal ancestor whose order of genes is the same in modern birds’ chromosomes.
“This remarkable finding shows the evolutionary stability of the order and orientation of genes on chromosomes over an extended evolutionary timeframe of more than 320 million years,” Lewin says. In contrast, regions between these conserved blocks contained more repetitive sequences and were more prone to breakages, rearrangements and sequence duplications, which are major drivers of genome evolution.
“Ancestral genome reconstructions are critical to interpreting where and why selective pressures vary across genomes. This study establishes a clear relationship between chromatin architecture, gene regulation and linkage conservation,” says Professor William Murphy, Texas A&M University, who was not an author on the paper. “This provides the foundation for assessing the role of natural selection in chromosome evolution across the mammalian tree of life.”
The scientists were able to follow the ancestral chromosomes forward in time from the common ancestor. They found that the rate of chromosome rearrangement differed between mammal lineages. For example, in the ruminant lineage (leading to modern cattle, sheep and deer) there was an acceleration in rearrangement 66 million years ago, when an asteroid impact killed off the dinosaurs and led to the rise of mammals.
“The results will help understanding the genetics behind adaptations that have allowed mammals to flourish on a changing planet over the last 180 million years,” explains co-author Dr Camilla Mazzoni, head of “Evolutionary and conservation genetics” at Berlin Center for Genomics in Biodiversity Research and Research Group Leader in Evolutionary and Conservation Genomics at the Department of Evolutionary Genetics at Leibniz-IZW.
#Nature
#2022 Science News#9-2022 Science News#acts of science#Earth Environment#earth science#Environment and Nature#everyday items#Nature Science#New#News Science Spies#Our Nature#planetary science#production line#sci_evergreen1#Science#Science Channel#science documentary#Science News#Science Spies#Science Spies News#September 2022 Science News#Space Physics & Nature#Space Science#Nature
0 notes
Text
West African plant knowledge and the origins of a unique species of African rice crop grown in the Americas; fugitive slaves and maroon communities maintaining knowledge of rice in liberated provision gardens; asserting the power and agency of West African knowledge against colonial erasure and obscurity; much more evidence supporting the “Black Rice thesis” has been discovered in the past 20 years; West African plant cultivation traditions in Suriname, Brazil, Colombia, Cuba, and South Carolina demonstrate strength of rice knowledge.
-------
In 2010 the American Historical Review published a special forum entitled “The Question of ‘Black Rice’”. The collection emerged from an animated debate among historians following the publication of Carney’s book nine years prior (Carney 2001). Black Rice argues that Africans had been growing a species of rice they domesticated independently [...] prior to the start of the transatlantic slave trade. With their knowledge and skills, and despite the constraints of enslavement, Africans introduced this culture into the plantations and societies of the New World. The more conventional belief that Europeans introduced rice into West Africa and then brought knowledge of its cultivation to the Americas, the book argues, is a primary fallacy [...]. At issue was a methodological question, namely what sources and approaches can count as evidence for reconstructing the cultural antecedents of New World developments. [...]
Black Rice broadened the sources of knowledge available to scholars [...]. By examining the transatlantic cultures and landscapes of rice, including its cultivation, milling, and cooking methods, the book analysed the Atlantic history of an important food crop from the perspective of those who grew it, both in Africa and its diaspora. This comparative approach thus draws attention to West Africa -- a region too often ignored in studies of the historical process shaping the environmental transformation of the Americas -- where a distinct species of rice, Oryza glaberrima, had been independently domesticated 3,500 years ago. Focusing on transatlantic flows of knowledge and cultural exchange, the book reconstructed the processes by which enslaved West Africans drove the development of rice landscapes and economies in the Americas using the ancestral skills and knowledge of their homelands. [...]
Despite opposition from some corners, studies of the African contributions to New World risiculture have advanced markedly in the years since the publication of Black Rice, building on its theories and methods and lending support to many of the book’s original findings.

In 2006, Dutch ethnobotanist Tinde van Andel discovered African rice, O. glaberrima, being cultivated in a maroon community in Suriname for use as ancestral offerings in the traditional Afro-Surinamese Winti religion. Applying historical linguistics and later rice genomics, her research built on the theories of African agency presented in Black Rice to explore how Suriname’s maroons have maintained ancestral connections by cultivating African varieties of rice introduced centuries earlier via the transatlantic slave trade [...]. The data suggest that the variety of African rice collected in Suriname likely originated in West Africa’s Guinea Plateau (van Andel et al. 2016a). Published in Nature Plants, the study became the first to use plant genomics to confirm the introduction of crops into the Americas by enslaved Africans [...].
It also shows how African rice endures in Neotropical America as a commemorative heritage plant, symbolising unbroken ancestral connections within a human history that could not be severed by centuries of slavery and racism.
Inspired by her findings with African rice, van Andel went on to examine the African origins of other basic foods produced by Suriname’s maroon communities -- from okra to yams, and from bananas to Bambara groundnuts -- which remain central to the diaspora’s cultural heritage (van Andel et al. 2014, 2016b).
The [...] research conducted by van Andel and her colleagues demonstrates that African rice was indeed introduced to the Americas and associates its cultivation with communities founded by fugitive slaves. Evidence from across the Atlantic World further suggests that similar processes of transatlantic diffusion replicated throughout the Western Hemisphere. In 2011, Carney located a botanical voucher of glaberrima rice in the Natural History Museum in London, originally collected in Matanzas, Cuba, in 1849 [...]. This specimen affirms that African rice was also cultivated in Cuba [...].Consequently, Carney’s original map in Black Rice showing the diffusion of African rice to New World slave societies can now be revised. [...]
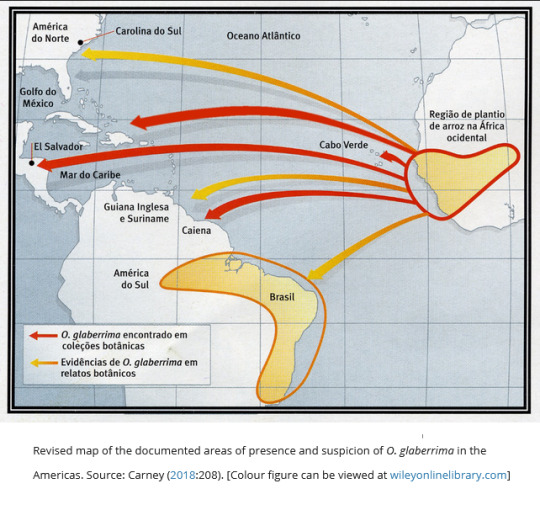
Recent work on the material culture surrounding the cultivation of rice in the Americas continues to bolster the Black Rice thesis. Historian Caroline Grego (2021) describes a mud-lifting shovel innovated from the kayendo -- a specialised farm implement developed in West African mangrove rice fields -- that was still in use on plantations in lowland South Carolina in the early 20th century. She blends analyses of historical photographs with field visits, oral histories, archives, and other sources to demonstrate a West African provenance for the shovels [...]. Grego’s research thus fills in more pieces to the complex puzzle of West African influences on the development of rice landscapes and economies in the Americas.
-------
Working beyond the colonial period, Eloisa Berman-Arévalo builds on Black Rice to analyse how communities in the Colombian Caribbean circumvented agrarian modernisation in the late 20th century through a collective rice harvest known as tongueo. Led by women, the tongueo “re-appropriates and re-signifies” the modern rice harvest as a joyous space of inter-generational Afrodiasporic cultural memory and knowledge transfer [...]. Two decades after publication, the theories and methods of Black Rice continue to influence histories and geographies of (post)colonialism and the African diaspora by clarifying the processes of exchange and innovation that coalesced in the early Atlantic World. [...]
In the many extant maroon communities scattered across northeastern South America, rice remains an esteemed food. Despite the vast distances that separate these communities, from Suriname to Brazil, descendants have passed down similar narratives about how they came to grow rice. Interviews with members of maroon communities (quilombolas) in the Brazilian states of Amapá, Pará, and Maranhão echo what the French botanist André Vaillant observed in French Guiana in 1936 (Carney 2004; Vaillant 1948). While collecting rice from maroon communities near the border with Suriname, Vaillant recorded the widespread belief that Black women introduced rice culture from Africa by hiding the grains in their hair. [...] These oral histories situate the origins of maroon rice culture around slave ships, excess provisions, and the agency of an enslaved African woman [...].
-------
Images, captions, and text published by: Case Watkins and Judith A. Carney. “Amplifying the Archive: Methodological Plurality and Geographies of the Black Atlantic.” Antipode. April 2022. [Italicized first paragraph/heading added by me.]
27 notes
·
View notes
Text
MYTHOLOGICAL
In populations across North Eurasia, from Indo-European groups in the far west to the Chukchi in the far east, as well as in the Americas, two styles of myth can be found with striking consistency; an "Earth Diver" creation myth, and a myth in which a monstrous dog guards the underworld, where getting past it acts a test for deceased souls.
The "Earth Diver" myth generally follows a plot where some animal/being is sent to dive to the bottom of the primordial ocean to collect a small amount of sand or mud, from which a supreme creator deity constructs the first dry land. This story, with minor variations, is found in the mythos of Northeast Asian, Siberian, and Eastern European populations such as the Samoyedic peoples, Turkic peoples, the Buryats, the Ainu, Chukchi, Yukaghir, Slavs, Lithuanians¹²³⁴, etc, as well as Native American groups; Blackfoot, Arapaho, Athabaskans, Cayuga, Nipmuc, Ojibway, Potawatomi, Mohawk⁵, etc.
The Afterlife-Dog-Guardian myth is found throughout Indo-European cultures; you are most likely familiar with Cerberus from Greek mythology. It's also found among the Chukchi and Tungusic peoples. There's remains of dogs that appear to have been ritualistically slaughtered at Mesolithic sites in Siberia, which may be related to this myth. In North America, the mythos of Siouan, Algonquian, and Iroquoian peoples also generally feature a fierce dog guarding the path to the afterlife; however, in their myths, the path is the Milky Way in the sky, rather than the underworld⁶.
Now, if this consistent continuum of distribution wasn't convincing enough, there's now also archaeogenetic evidence that further implies these two myths have a common origin (which would make them among the oldest reconstructable stories we know of.) Genetic analysis infers the existence of a Paleolithic/Mesolithic grouping of people in North Eurasia aptly known as the Ancient North Eurasians. The thread that defines the ANE is close relation to the 24,000 year old Mal'ta Boy remains from the Lake Baikal region⁷. The Ancient North Eurasians were an extremely prolific and important population; to the East, they've contributed ancestry to modern Siberian populations (and to a lesser extent, East Asians), and a large amount to Native Americans; 40-50% of Native American DNA is ANE in origin⁷. To the west, they mixed with various West Eurasian hunter-gatherers to form Mesolithic lineages such as the Caucasus Hunter Gatherers, Eastern European Hunter Gatherers, West Siberian Hunter Gatherers, and Scandinavian Hunter Gatherers. Proto-Indo-Europeans descended from a mixture of Eastern European Hunter Gatherers (70-75% ANE, 30-25% Western European Hunter Gatherer) and Caucasus Hunter Gatherers⁸ (45-62% Dzudzuana/Archaic Caucasus HG, 55-38% ANE⁹). This shared ancestry lends credence to the idea that this pattern of myths could have a common origin in North Asia predating the migration of people to the Americas.
Furthermore, very recent research (a few months ago) suggests that the domestication of the dog occurred in Siberia around 23,000 years ago, where, from there, they spread throughout Eurasia and into the Americas; the people responsible for the domestication are identified in the initial study as "Ancient North Siberians"¹⁰ , though they were described as being associated with the Mal'ta remains and are thus synonymous with Ancient North Eurasians. Not necessarily strong evidence in its own right that the Afterlife Dog myth comes from them, but I definitely think it's worth mentioning. There's also rituals consistently associating dogs with healing and protection through their perceived ability to absorb illness found in ANE-descended populations such as the Hittites, ancient Greeks, ancient Italics, Turkic peoples, Babylonians (Mesopotamia is adjacent to where Neolithic Iranians lived, who also had a large amount of ANE ancestry), and possibly the ancient Botai culture⁶.
(And just as a fun fact, blonde hair among West Eurasians is believed to have originated with the ANE; the earliest person known to have been genetically predisposed to blonde hair was the ~17,000 year old ANE-descended Afontova Gora 3 girl from central Siberia.)
CULTURAL
Broadly similar religious practices that can be described as shamanism are mostly distributed among East Asians (especially Siberians) and, quite famously, Native Americans¹². Uralic peoples, who ultimately originate from east Siberia¹³, brought it to Europe. The fact that shamanism is found in many different East Asian populations, including ones with little ANE ancestry in southern China, east India, and Southeast Asia, but is/was essentially absent in West Eurasia (as far as we know), suggests that the practice has an ancient East Asian origin, and that Native Americans picked it up from their East Asian ancestral component (40-50% of their DNA from ANE, 60-50% from Ancient East Asian)
Of course, shamanistic practices are also present in cultures from Sub Saharan Africa, and among Australian Aboriginals, so as a whole the concept obviously emerged multiple times, but the particularly strong concentration of it among the deeply related peoples of East Asia, Siberia, and Native America might indicate that theirs does have some sort of common heritage.
LINGUISTIC
In 2010, linguist Edward Vadja published The Dene–Yeniseian Connection, the culmination of years of his research, wherein he proposes that the Yeniseian language family of Central Siberia, and the Na-Dene language family of North America (which includes such languages as Navajo and Apache), are related¹⁴. This paper was actually well received by a number of respected linguists, the first theory connecting Old World and New World languages to receive this treatment. In a 2012 presentation, Vadja bolstered his theory with further comparative linguistic evidence, as well as non-linguistic evidence; the most compelling example of the latter being the fact that haplogroup Q1, which is very common throughout indigenous populations of the Americas , is found at a rate of nearly 90% among the Yeniseian Ket people (and at a rate of 65% in their neighbors, the Samoyedic-speaking Selkup, who have long intermixed with Yeniseians.) The Navajo have this haplogroup at a rate of 92%. The Ket people also have among the highest amount of ANE-derived ancestry out of any modern day population, similar to Native Americans¹⁵.
A possible scenario explaining the proliferation of the family: when the ancestors of most other Native Americans migrated over into the New World, a subset of them remained behind in Beringia for several millennia. Eventually, as Beringia began to flood ~10,000 years ago, the group split, with some going back into Asia to become the Yeniseians while others finally moved into America to become the Na-Dene. While this is certainly much older than the dating for most other established language families, it is not implausible; Proto-Afro-Asiatic, for example, is routinely dated to well over 10,000 years ago. However, Vadja himself postulated that Na-Dene might have arrived in America through a distinct migration which postdated the initial peopling of the continents.¹⁶
Today, Dene-Yeniseian is still not a universally accepted, firmly established language family, but its not widely rejected either; it remains an open, favorable possibility while further research is done. For the sake of fairness, here is an example of a critical review which doubts the theory as Vadja proposed it: http://www2.hawaii.edu/~lylecamp/Campbell%20Yeniseian%20NaDene%20review%2011-2-10.pdf
SOURCES
Sacred narrative, readings in the theory of myth, Alan Dundes, page 168-70
Earth-Diver Myth (А812) in northern Eurasia and North America: twenty years later, Vladimir Napolskikh
A Dictionary of Asian Mythology, David Adams Leeming, page 55
The Motifs of creating the world in the Lithuanian narrative folklore, Nijolė Laurinkienė
http://www.native-languages.org/earthdiver.html
Tracing the Indo-Europeans: New evidence from archaeology and historical linguistics, David W. Anthony & Dorcas R. Brown, page 104-105
Upper Palaeolithic Siberian Genome Reveals Dual Ancestry of Native Americans, Pontus Skoglund & Maanasa Raghavan
Archaeology, Genetics, and Language in the Steppes: A Comment on Bomhard, David Anthony
Paleolithic DNA from the Caucasus reveals core of West Eurasian ancestry, Josif Lazaridis, Anna Belfer-Cohen, Swapan Mallick, Nick Patterson, Olivia Cheronet,Nadin Rohland Guy Bar-Oz, Ofer Bar-Yosef, Nino Jakeli, Eliso Kvavadze, David Lordkipanidze, Zinovi Matzkevich, Tengiz Meshveliani, Brendan J. Culleton, Douglas J. Kennett, Ron Pinhasi, David Reich
Dog domestication and the dual dispersal of people and dogs into the Americas, Angela R. Perri , Tatiana R. Feuerborn, Laurent A. F. Frantz, Greger Larson , Ripan S. Malhi, David J. Meltzer, Kelsey E. Witt
The genomic history of southeastern Europe, David Reich + many many more names
https://upload.wikimedia.org/wikipedia/commons/0/0c/Map_of_Shamanism_across_the_world.svg
The Arrival of Siberian Ancestry Connecting the Eastern Baltic to Uralic Speakers Further East, Lehti Saag
Reviewed Work: The Dene–Yeniseian Connection by James Kari and Ben A. Potter
Genomic study of the Ket: a Paleo-Eskimo-related ethnic group with significant ancient North Eurasian ancestry, Pavel Flegontov
DENE-YENISEIAN LANGUAGE FAMILY: EVIDENCE FOR A BACK-MIGRATION TO THE OLD WORLD? German Dziebel
25 notes
·
View notes
Text
Here’s the most exciting thing I’ve learned this month: the concrete information theoretic limit to how far back you can learn about ancestral human populations using the DNA of modern humans!
I’m confused by the explanation of ‘splices’ and cannot appreciate this fully, but what I’m getting is still very cool!
Females create an average of about forty-five new splices when producing eggs, while males create about twenty-six splices when producing sperm, for a total of about seventy-one new splices per generation. ...
Any person’s genome is derived from 47 stretches of DNA corresponding to the chromosomes transmitted by mother and father plus mitochondrial DNA. One generation back, a person’s genome is derived from about 118 (47 plus 71) stretches of DNA transmitted by his or her parents. Two generations back, the number of ancestral stretches of DNA grows to around 189 (47 plus 71 plus another 71) transmitted by four grandparents. Look even further back in time, and the additional increase in ancestral stretches of DNA every generation is rapidly overtaken by the doubling of ancestors. Ten generations back, for example, the number of ancestral stretches of DNA is around 757 but the number of ancestors is 1,024, guaranteeing that each person has several hundred ancestors from whom he or she has received no DNA whatsoever. Twenty generations in the past, the number of ancestors is almost a thousand times greater than the number of ancestral stretches of DNA in a person’s genome, so it is a certainty that each person has not inherited any DNA from the great majority of his or her actual ancestors.
These calculations mean that a person’s genealogy, as reconstructed from historical records, is not the same as his or her genetic inheritance. The Bible and the chronicles of royal families record who begat whom over dozens of generations. Yet even if the genealogies are accurate, Queen Elizabeth II of England almost certainly inherited no DNA from William of Normandy, who conquered England in 1066 and who is believed to be her ancestor twenty-four generations back in time. This does not mean that Queen Elizabeth II did not inherit DNA from ancestors that far back, just that it is expected that only about 1,751 of her 16,777,216 twenty-fourth-degree genealogical ancestors contributed any DNA to her.

Figure 4. The number of ancestors you have doubles every generation back in time. However, the number of stretches of DNA that contributed to you increases by only around seventy-one per generation. This means that if you go back eight or more generations, it is almost certain that you will have some ancestors whose DNA did not get passed down to you. Go back fifteen generations and the probability that any one ancestor contributed directly to your DNA becomes exceedingly small.
Going back deeper in time, a person’s genome gets scattered into more and more ancestral stretches of DNA spread over ever-larger numbers of ancestors. Tracing back fifty thousand years in the past, our genome is scattered into more than one hundred thousand ancestral stretches of DNA, greater than the number of people who lived in any population at that time, so we inherit DNA from nearly everyone in our ancestral population who had a substantial number of offspring at times that remote in the past.
There is a limit, though, to the information that comparison of genome sequences provides about deep time. At each place in the genome, if we trace back our lineages far enough into the past, we reach a point where everyone descends from the same ancestor, beyond which it becomes impossible to obtain any information about deeper time from comparison of the DNA sequences of people living today. From this perspective, the common ancestor at each point in the genome is like a black hole in astrophysics, from which no information about deeper time can escape. For mitochondrial DNA this black hole occurs around 160,000 years ago, the date of “Mitochondrial Eve.” For the great majority of the rest of the genome the black hole occurs between five million and one million years ago, and thus the rest of the genome can provide information about far deeper time than is accessible through analysis of mitochondrial DNA. Beyond this, everything goes dark.
325 notes
·
View notes
Text
Fwd: Conference: Montreal.QuestforOrthologs.Jul17-18.DeadlineMay14
Begin forwarded message:
> From: [email protected]
> Subject: Conference: Montreal.QuestforOrthologs.Jul17-18.DeadlineMay14
> Date: 4 May 2024 at 05:26:56 BST
> To: [email protected]
>
>
> PLEASE FORWARD THIS ANNOUNCEMENT!
>
> Quest for Orthologs meeting (QfO8) - July 17-18, 2024 - Montr�al,
> QC, Canada.
>
> LAST CALL : abstract submissions for Talks and/or Posters.
>
> DEADLINE EXTENSION TO : May 14, 2024
>
> Webpage:
> https://ift.tt/2ZQDByi
>
> Confirmed Invited Speakers : Fiona Brinkman (Simon Fraser University),
> Elena Kuzmin (Concordia University), Markus Hecker (University of
> Saskatchewan), Christine Orengo (University College London).
>
> Scope: The QfO Consortium addresses a key challenge in molecular
> evolutionary biology and strives to develop large-scale methods for
> inferring orthology among fully sequenced genomes, and to reconstruct the
> complement of protein-coding genes in common ancestral genomes across the
> tree of life. The QFO meetings aim to bring together disparate groups
> around the world that work in the field to accelerate progress through
> sharing of ideas, methods and research findings. The particular aim of
> this QfO8 meeting is to gather an even more diverse set of end-users and
> to connect different research communities that are rooted in orthology.
>
> Travel and Caregiver Fellowships: Authors of accepted talks/posters
> will be eligible for travel and caregiver fellowships to attend the
> conference. The procedure for application will be communicated to the
> relevant people in the talk/poster acceptance email.
>
> ISCB members can receive a 15% discount on registration rates.
>
> Further information on abstract submission deadline and guidelines can
> be found here: https://ift.tt/WGTmF1N
>
> PLEASE FORWARD THIS ANNOUNCEMENT!
>
>
> A�da Ouangraoua
0 notes
Link
By Lizzie Wade
Apr. 18, 2018, 3:00 PM
Today, the population of South Asia is divided into dozens of ethnic, linguistic, and religious groups that live side by side—but not always in harmony. A contentious border separates India and Pakistan; political movements draw stark lines between India’s Muslim and Hindu populations. Groups don’t mix much, as people tend to marry those who share their ethnicity and tongue.
Now, a study of the first ancient DNA recovered from South Asia shows that populations there mingled repeatedly thousands of years ago. Nearly all of the Indian subcontinent’s ethnic and linguistic groups are the product of three ancient Eurasian populations who met and mixed: local hunter-gatherers, Middle Eastern farmers, and Central Asian herders. Three similar groups also mingled in ancient Europe, giving the two subcontinents surprisingly parallel histories.
The study, presented here last week at the meeting of the American Association of Physical Anthropologists and in a preprint on the bioRxiv server, sheds light on where these populations came from and when they arrived in South Asia. It also strengthens the claim that Proto-Indo-European (PIE)—the ancestral language that gave rise to modern languages from English to Russian to Hindi—originated on the steppes of Asia.
“It’s first-rate work,” says Partha Majumder, a geneticist at the National Institute of Biomedical Genomics in Kalyani, India. He found hints of similar genetic patterns in his previous studies, but the addition of ancient DNA makes the new conclusions stronger, he says. “It’s absolutely stunning.”
Priya Moorjani, a geneticist at the University of California, Berkeley, studies how South Asian populations relate to each other and to others around the world. In previous work, she analyzed the genomes of nearly 600 modern Indians and Pakistanis from 73 ethnolinguistic groups in South Asia. Her team found that almost all people living in India today carry ancestry from two ancient populations: Ancestral North Indians, who were more related to people from Central Asia, the Middle East, the Caucasus, and Europe; and Ancestral South Indians, who were more related to indigenous groups living in the subcontinent today. But without DNA from ancient people, Moorjani couldn’t be sure who gave rise to those ancestral populations, or when.
Moorjani, David Reich of Harvard University, and Kumarasamy Thangaraj of the Centre for Cellular and Molecular Biology in Hyderabad, India, spent years searching for ancient DNA in South Asia, where hot climates might degrade it. Finally, their team recovered and analyzed ancient genomes from 65 individuals who lived in northern Pakistan between 1200 B.C.E. and 1 C.E. They also analyzed 132 ancient genomes from Iran and southern Central Asia, and 165 from the steppes of Kazakhstan and Russia, and compared them with published ancient and modern genomes. These data allowed them to reconstruct when different populations arrived in South Asia and how they interacted.
Between 4700 and 3000 B.C.E., farmers from Iran mixed with hunter-gatherers indigenous to South Asia, Moorjani said. This combination of ancestries was found in the DNA of skeletal remains from sites in Turkmenistan and Iran known to have been in contact with the Indus Valley civilization, which thrived in Pakistan and northwest India starting around 3300 B.C.E. The researchers dub this population “Indus periphery.” The 65 ancient people from Pakistan also show this combination, although they all lived after the Indus civilization declined. The researchers suspect that “Indus periphery” people actually may have been the founders of Indus society, although without ancient DNA from Indus Valley burials, they can’t be sure.
Still, Moorjani’s team sees this ancient mixture of Iranian farmers and South Asian hunter-gatherers all over South Asia today. As the Indus Valley civilization declined after 1300 B.C.E., some Indus periphery individuals moved south to mix with indigenous populations there, forming the Ancestral South Indian population, which today is more prominent in people who speak Dravidian languages such as Tamil and Kannada, and in those belonging to lower castes.
Meanwhile, herders from the Eurasian steppe moved into the northern part of the subcontinent and mixed with Indus periphery people still there, forming the Ancestral North Indian population. Today, people who belong to higher castes and those who speak Indo-European languages such as Hindi and Urdu tend to have more of this ancestry. Shortly after, these two already mixed groups mixed with each other, giving rise to the populations living in India today.
“Strikingly, this is very similar to the pattern we see in Europe,” Moorjani said. Around 7000 B.C.E., agriculture spread into both Europe and South Asia with farmers from Anatolia and Iran, respectively, who each mixed with local hunter-gatherer populations. After about 3000 B.C.E., Yamnaya pastoralists from the Central Asian steppe swept both east and west, into Europe and South Asia, bringing the wheel and perhaps cannabis.
Earlier genetic work had linked the arrival of these herders to the spread of Indo-European languages in Europe. But other researchers, including archaeologist Colin Renfrew of the University of Cambridge in the United Kingdom, had argued that the earlier Anatolian farmers were the original PIE speakers. The new data “make a strong case” for the Yamnaya as carriers of Indo-European languages, Renfrew says. But he still thinks Anatolian farmers could have spoken the earliest language in that family.
7 notes
·
View notes