#biorxiv
Explore tagged Tumblr posts
Text


@shiftythrifting
I can relate to how this mug came into existence.
308 notes
·
View notes
Text
I guess general PSA: if a new scientific paper/study is being passed around and it is from arxiv.org, biorxiv.org, medrxiv.org, etc. that means it is a preprint, and has not yet been peer reviewed. that doesn't mean it's wrong, but it does mean that it has no experts to vouch for it who didn't work directly on it.
even a peer-reviewed study should be looked at with skepticism, but a study that hasn't been peer-reviewed should be looked at with double that.
16 notes
·
View notes
Text

A New Study Finds That Domestic Cats Traveled the Silk Road to China About 1,400 Years Ago
The animals were likely gifted to some elites, then spread throughout the region.
The modern world’s pet cats are products of a long process of domestication and trade. It began some 10,000 years ago in modern-day Turkey, when Anatolians tamed and befriended Near Eastern wildcats. About 7,000 years later, these companion cats spread to Europe through trade. But scientists have long wondered when and how feline friends arrived in China—where they’re currently the most popular urban pet. Now, using genetic testing, researchers have discovered that pet cats likely arrived in China around 600 C.E.—over 1,500 years after their introduction to Europe. According to a study recently published on the preprint server bioRxiv, cats were one of the many assets that traveled east on the famous Silk Road, the lengthy trading network that connected Asia with Europe between the second century B.C.E. and the 15th century C.E. Per the study, China’s oldest known domestic cat lived in the central province of Shaanxi between 706 and 883, during the Tang dynasty. It likely sported a short, all-white or partly-white coat—like most Chinese house cats today—and a long tail, and its ancestors probably came from Kazakhstan...
Read more: https://www.smithsonianmag.com/smart-news/a-new-study-finds-that-domestic-cats-traveled-the-silk-road-to-china-about-1400-years-ago-180986206/
131 notes
·
View notes
Text
A small team of biologists at the University of Bristol has found that black garden ants modify the physical structure of their nests to mitigate infection spread. The group has written a paper describing the experiments they conducted with black garden ants and fungal infections in their lab and posted it on the bioRxiv preprint server. Prior research has shown that some animals change their behavior to avoid spreading infections, whether they be viral, bacterial or fungal. Among those, only humans have been found to alter their surroundings as a way to further protect themselves—people might close off parts of their house, for example, or establish quarantine zones within hospital areas.
Continue Reading.
166 notes
·
View notes
Text
We desperately need a vaccine that works for all variants.

164 notes
·
View notes
Text
Also preserved in our archive
By Dr. Sushama R. Chaphalkar, PhD.
In a recent research paper posted to the bioRxiv preprint* server, researchers in the United States investigated the potential effect of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection on cholesterol metabolism, focusing on the role of the viral protein open reading frame 3a (ORF3a).
They found that SARS-CoV-2 causes cholesterol sequestration in lysosomes via the ORF3a protein, which disrupts protein trafficking and reduces the levels of bis(monoacylglycero)phosphate (BMP) in the cell, enhancing viral survival.
Background Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, disrupts lipid metabolism, particularly cholesterol homeostasis, which can persist during and after infection. This is linked to disease severity and long-term complications like dyslipidemia and cardiovascular diseases.
Cholesterol is crucial for cellular function and is primarily transported through lysosomes, where proteins like Niemann-Pick C1 and C2 (NPC1 and NPC2) facilitate its release. SARS-CoV-2 may exploit plasma membrane cholesterol to enhance infectivity.
Disruptions in the lysosomal cholesterol pathway can cause cholesterol buildup, impairing cellular functions, and viruses like Ebola are known to hijack this mechanism. Notably, BMP plays a dual role: it aids in cholesterol transport and contributes to viral infection by promoting viral fusion with lysosomal membranes.
In the present study, researchers investigated the potential impact of SARS-CoV-2 infection on cholesterol transport in cells, focusing on the role of the viral protein ORF3a.
About the Study A variety of experimental techniques were employed, including culturing A549, HeLa, and Vero E6 cells, followed by SARS-CoV-2 infection at different multiplicities of infection. SARS-CoV-2 ORF3a-VPS39 interaction was studied using mutations at key residues (notably W193 and Y184, which were identified as critical for this interaction). Immunofluorescence, filipin staining, and confocal microscopy were used to assess cholesterol localization and vesicular dynamics, while high-content imaging quantified cell-specific responses.
Cholesterol levels were measured using gas chromatography-mass spectrometry (GC-MS), and lipid species were analyzed through shotgun lipidomics. For further protein analysis, western blotting was performed to detect secreted NPC2 and cathepsin D, along with cell lysates. Data were analyzed using ImageJ and Prism 9, and statistical significance was determined by t-tests or analysis of variance.
Results and Discussion SARS-CoV-2 infection was found to increase filipin-positive puncta in lysosomes of A549-hACE2 and Vero E6 cells, indicating altered cholesterol distribution, especially in lysosomes, without affecting total cholesterol levels. Among the 28 viral proteins tested, ORF3a showed the strongest increase in filipin puncta, suggesting significant lysosomal cholesterol sequestration.
Notably, SARS-CoV-2 ORF3a localized to lysosomes and caused them to swell, whereas SARS-CoV ORF3a did not induce such effects, highlighting a distinct pathogenic strategy unique to SARS-CoV-2.
ORF3a was found to interact with VPS39, a key component of the HOPS complex involved in cholesterol egress from lysosomes. Key residues W193 and Y184 were shown to form a hydrophobic binding interface critical for this interaction, distinguishing SARS-CoV-2 ORF3a from its SARS-CoV counterpart. Mutations at W193 and Y184 disrupted this interaction, while S171 and H182 had no significant effect.
SARS-CoV-2 ORF3a expression was shown to cause cholesterol accumulation in lysosomes, which was reduced by the W193A mutation. It also led to the mislocalization of NPC2 and increased its secretion, indicating disrupted NPC2 trafficking, likely due to interference with TGN-to-endosome transport. Additionally, BMP levels were significantly reduced in infected cells, which likely exacerbates lysosomal cholesterol sequestration.
In SARS-CoV-2-infected Vero E6 cells, BMP levels were found to decrease at 12 hours post-infection, coinciding with increased cholesterol at 18 hours. In HeLa-Flp-In cells, SARS-CoV-2 ORF3a was found to reduce BMP levels by 20%, with partial rescue in the W193A mutant. Lipidomics confirmed this reduction, correlating BMP loss with cholesterol accumulation and suggesting BMP reduction may contribute to cholesterol sequestration.
SARS-CoV-2 may reduce plasma membrane cholesterol to limit secondary infections, as shown by decreased SARS-CoV-2 infection in NPC1 inhibitor-treated cells. This supports the hypothesis that the virus manipulates cholesterol distribution to optimize replication conditions. Interestingly, the virus also appears to reduce its own infectivity within a single cell, suggesting a self-regulating mechanism to prevent viral overload and ensure broader host-level spread.
Conclusion In conclusion, a novel mechanism by which SARS-CoV-2 disrupts host cell lipid metabolism, specifically through cholesterol sequestration in lysosomes, has been elucidated. By uncovering the specific interaction between the viral protein ORF3a and host protein VPS39, the study highlights a critical role of lysosomal cholesterol trafficking disruption in SARS-CoV-2 pathogenesis.
This discovery opens potential therapeutic avenues to target lipid dysregulation in COVID-19, which could help mitigate both the disease's immediate and long-term metabolic consequences, including dyslipidemia and cardiovascular complications.
Journal reference: Preliminary scientific report. Manipulation of Host Cholesterol by SARS-CoV-2. Aliza Doyle et al., bioRxiv, 2024.11.13.623299 (2024), DOI: 10.1101/2024.11.13.623299,
Study Link: www.biorxiv.org/content/10.1101/2024.11.13.623299v1
#mask up#public health#wear a mask#pandemic#covid#wear a respirator#covid 19#still coviding#coronavirus#sars cov 2
91 notes
·
View notes
Text
General announcement: we've made exceptions before but we don't usually publish submissions from preprint servers like arXiv and bioRxiv. These websites are archives for manuscripts at various stages of completion, not scientific journals, so the manuscripts they host often are not yet peer reviewed.
Our goal is to spotlight jokes, mistakes, or other oddities in scientific papers that made it through the peer review and editing process, so manuscripts at an unreviewed preprint stage are technically outside the aims and scope of the blog.
198 notes
·
View notes
Text
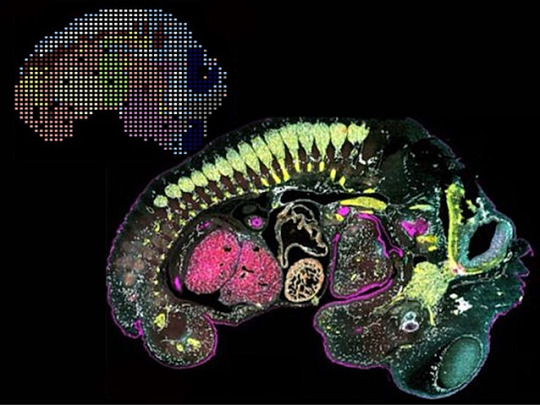
Seen Two Ways
Genes and proteins are two sides of the same coin. Genes are sections of DNA that contain the instructions for producing proteins, which themselves play countless fundamental roles in the body – including regulating DNA expression. Researchers have techniques to measure gene expression or protein levels within cells and tissues, but a new approach marries the two to show a more rounded picture of a single specimen, such as the mouse embryo pictured. Clusters of gene expression patterns (top left) can be compared with the location and amounts of proteins (bottom, with colours indicating different proteins). Connecting gene activity and the precise location of protein function provides a level of insight that could answer important questions about health and disease, such as why some regions of tumours express genes differently, how protein imbalances in the brain are linked to neurodegenerative disorders, or how immune cells interact with tissues during inflammation.
Written by Anthony Lewis
Image from work by Archibald Enninful and Zhaojun Zhang, and colleagues
Department of Biomedical Engineering, Yale University, New Haven, CT, USA
Image contributed by the authors under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence
Published in bioRxiv, February 2024 (not peer reviewed)
You can also follow BPoD on Instagram, Twitter, Facebook and Bluesky
#science#biomedicine#immunofluorescence#biology#cancer#genes#inflammation#neurodegenerative diseases
11 notes
·
View notes
Text

María Branyas Morera, quien falleció en agosto de 2024 a los 117 años y 168 días. No solo ostentaba un récord Guinness como la persona más longeva del mundo, sino que también formó parte de un estudio destinado a desentrañar el misterio de su sorprendente genética. Publicado en BioRxiv, este estudio podría aportar claves fundamentales para comprender la longevidad extrema y la resistencia a enfermedades asociadas con la vejez.
Lee la nota completa ingresando al siguiente link:
4 notes
·
View notes
Text





We never tire of celebrating preprints! Cheers to lettuce, Prf and friends! Kudos Daniel, Pai, AmirAli and Team!
Luedke, D., Pai, H., Toghani A., Harant, A., Wu, C.-H., and Kamoun, S. 2025. The autoactivity of tomato helper NLR immune proteins of the NRC clade is unaltered in prf mutants of Nicotiana benthamiana. bioRxiv, doi: https://doi.org/10.1101/2025.03.11.642614. OpenPlantNLR dataset
Pai, H., Sakai, T., Posbeyikian, A., Frijters, R., Sugihara, Y., Contreras, M.P., Kourelis, J., Adachi, H., Kamoun, S., and Toghani A. 2025. A hierarchical immune receptor network in lettuce reveals contrasting patterns of evolution in sensor and helper NLRs. bioRxiv, doi: https://doi.org/10.1101/2025.02.25.639832. OpenPlantNLR dataset
2 notes
·
View notes
Text
De-extinction startup Colossal Biosciences wants to bring back the woolly mammoth. Well, not the woolly mammoth exactly, but an Asian elephant gene-edited to give it the fuzzy hair and layer of blubber that allowed its close relative to thrive in sub-zero environments.
To get to these so-called “functional mammoths,” Colossal’s scientists need to solve a whole bunch of challenges: making the right genetic tweaks, growing edited cells into fully formed baby functional mammoths, and finding a space where these animals can thrive. It’s a long, uncertain road, but the startup has just announced a small breakthrough that should ease some of the way forward.
Scientists at Colossal have managed to reprogram Asian elephant cells into an embryonic-like state that can give rise to every other cell type. This opens up a path to creating elephant sperm and eggs in the lab and being able to test gene edits without having to frequently take tissue samples from living elephants. The research, which hasn’t yet been released in a peer-reviewed scientific journal, will be published on the preprint server Biorxiv.
There are only around 30,000 to 50,000 Asian elephants in the wild, so access to these animals—and particularly their sperm and eggs—is extremely limited. Yet Colossal needs these cells if they’re going to figure out how to bring their functional mammoths to life. “With so few fertile female elephants, we really don’t want to interfere with their reproduction at all. We want to do it independently,” says George Church, a Harvard geneticist and Colossal cofounder.
The cells that Colossal created are called induced pluripotent stem cells (iPSCs), and they behave a lot like the stems cells found in an embryo. Embryonic stem cells have the ability to give rise to all kinds of different cell types that make up organisms—a quality that scientists call pluripotency. Most cells, however, lose this ability as the organism develops. Human skin, for instance, can’t spontaneously turn into muscle or cells that line the inside of the intestine.
In 2006, the Japanese scientist Shinya Yamanaka showed it was possible to take mature cells and turn them back into a pluripotent state. Yamanaka’s research was in mice cells, but later scientists followed up by deriving iPSCs for lots of different species, including humans, horses, pigs, cattle, monkeys, and the northern white rhino—a functionally extinct subspecies with only two individuals, both females, remaining in the wild.
Reprogramming Asian elephant cells into iPSCs proved trickier than with other species, says Eriona Hysolli, head of biological sciences at Colossal. As with other species, the scientists reprogrammed the elephant cells by exposing them to a series of different chemicals and then adding proteins called transcription factors that turn on particular genes to change how the cells functions. The whole process took two months, which is much longer than the 5 to 10 days it takes to create mouse iPSCs or the three weeks for human iPSCs.
This difficulty might have to do with the unique biology of elephants, says Vincent Lynch, a developmental biologist at the University at Buffalo in New York who wasn’t involved in the Colossal study. Elephants are the classic example of Peto’s paradox—the idea that very large animals have unusually low rates of cancer given their size. Since cancer can be caused by genetic mutations that accumulate as cells divide, you’d expect that animals with 100 times more cells than humans would have a much higher risk of cancer.
But elephants have cancer rates even lower than humans—a surprising fact given their vast size. One hypothesis for elephants’ cancer-defying biology is that they carry lots of copies of a tumor-suppressing gene called P53. Humans, on the other hand, only have one copy of this gene.
P53 is good for elephant health, but it could be the reason that up until now scientists have struggled to create iPSCs from elephant cells, Lynch says. One way the gene seems to work is by stopping cells from entering a state where they can duplicate indefinitely, which is one of the key features of iPSCs.
Hysolli says that she’d like to reduce the time it takes to create elephant iPSCs, and refine the process so the Colossal team can produce them at a greater scale. The iPSCs will be particularly useful if Colossal’s scientists can turn them into sperm and egg cells, something that Hysolli’s team is already working on. Since there is a relatively limited supply of elephant eggs and sperm, one problem facing the de-extinction project is getting enough genetic diversity to support a population of functional mammoths—develop them from too few individuals, and you risk the negative effects of inbreeding. Being able to create sperm and egg cells in the lab should help with that, Church says.
These cells could also be useful for conservation work, Hysolli says. Colossal has partnered with researchers working on elephant endotheliotropic herpes virus (EEHV), a leading cause of death for young Asian elephants. The iPSCs could be a good way to figure out how the virus infects different cell types. The cells will also be useful for testing whether Colossal’s edits to produce mammoth-like fur and fat layers are working as scientists hope.
“I have no doubt that given enough time and money they will overcome the technical challenges of making a woolly-mammoth-looking elephant,” says Lynch. But he’s less convinced of the ecological benefits of de-extinction. The startup intends to introduce the elephant-mammoth hybrids into the wild to re-create the role once played by the mammoth in the Arctic ecosystem, grazing the land and trampling snow cover, potentially decelerating the melting of permafrost.
“How many hairy Asian elephants do you need to make that work?” Lynch asks. Whether there really is a niche for edited elephants in the Arctic 4,000 years after mammoths last roamed the area is a question that conservationists are still grappling with. Sure, scientists might be able to create mammoth-like Asian elephants, but whether we should is open to much debate.
Colossal’s scientists will be glad if they get to that point. Although they have elephant iPSCs, much of the work of creating elephant-mammoth hybrids is ahead of them. They must figure out how to create elephant sperm and egg cells, master the right edits to tweak their elephants, and take their creation through the 22-month Asian elephant gestation period. And then they have to do it enough times to build a population that can actually deliver on some of their ecological aims.
“It feels very significant,” Church says of the iPSC breakthrough. “This is a very big deal.” If Colossal is going to deliver on its de-extinction mission, then there will be many other moments like this ahead.
6 notes
·
View notes
Text
Excerpt:
Our mouths and guts are teeming with mysterious somethings that are unknown to science, new research suggests. A team says they’ve discovered distinct virus-like structures hanging out among the bacteria that live in our bodies. The researchers have coined these structures “obelisks,” and they might further redefine what it means to be a living thing. ...
“As such, obelisks comprise a class of diverse RNAs that have colonized, and gone unnoticed in, human, and global microbiomes,” the authors wrote in their preprint paper, released on bioRxiv this month.
7 notes
·
View notes
Text
The constant ebb and flow of hormones that guide the menstrual cycle don't just affect reproductive anatomy. They also reshape the brain, and a new study has given us insight into how this happens. Led by neuroscientists Elizabeth Rizor and Viktoriya Babenko of the University of California Santa Barbara, a team of researchers tracked 30 women who menstruate over their cycles, documenting in detail the structural changes that take place in the brain as hormonal profiles fluctuate. The results, which are yet to be peer-reviewed but can be found on preprint server bioRxiv, suggest that structural changes in the brain during menstruation may not be limited to those regions associated with the menstrual cycle. "These results are the first to report simultaneous brain-wide changes in human white matter microstructure and cortical thickness coinciding with menstrual cycle-driven hormone rhythms," the researchers write.
Continue Reading.
733 notes
·
View notes
Text
In vivo Antibody Painting for Next Generation Weight Loss Drugs
bioRxiv: http://dlvr.it/TCJHLd
2 notes
·
View notes
Text
Also preserved in our archive
By Vijay Kumar Malesu
In a recent pre-print study posted to bioRxiv*, a team of researchers investigated the predictive role of gut microbiome composition during acute Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection in the development of Long Coronavirus Disease (Long COVID) (LC) and its association with clinical variables and symptom clusters.
Background LC affects 10–30% of non-hospitalized individuals infected with SARS-CoV-2, leading to significant morbidity, workforce loss, and an economic impact of $3.7 trillion in the United States (U.S.).
Symptoms span cardiovascular, gastrointestinal, cognitive, and neurological issues, resembling myalgic encephalomyelitis and other post-infectious syndromes. Proposed mechanisms include immune dysregulation, neuroinflammation, viral persistence, and coagulation abnormalities, with emerging evidence implicating the gut microbiome in LC pathogenesis.
Current studies focus on hospitalized patients, limiting generalizability to milder cases. Further research is needed to explore microbiome-driven predictors in outpatient populations, enabling targeted diagnostics and therapies for LC’s heterogeneous and complex presentation.
About the study The study was approved by the Mayo Clinic Institutional Review Board and recruited adults aged 18 years or older who underwent SARS-CoV-2 testing at Mayo Clinic locations in Minnesota, Florida, and Arizona from October 2020 to September 2021. Participants were identified through electronic health record (EHR) reviews filtered by SARS-CoV-2 testing schedules.
Eligible individuals were contacted via email, and informed consent was obtained. Of the 1,061 participants initially recruited, 242 were excluded due to incomplete data, failed sequencing, or other issues. The final cohort included 799 participants (380 SARS-CoV-2-positive and 419 SARS-CoV-2-negative), providing 947 stool samples.
Stool samples were collected at two-time points: weeks 0–2 and weeks 3–5 after testing. Samples were shipped in frozen gel packs via overnight courier and stored at −80°C for downstream analyses. Microbial deoxyribonucleic acid (DNA) was extracted using Qiagen kits, and metagenomic sequencing was performed targeting 8 million reads per sample.
Taxonomic profiling was conducted using Kraken2, and functional profiling was performed using the Human Microbiome Project Unified Metabolic Analysis Network (HUMAnN3).
Stool calprotectin levels were measured using enzyme-linked immunosorbent assay (ELISA), and SARS-CoV-2 ribonucleic acid (RNA) was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Clinical data, including demographics, comorbidities, medications, and symptom persistence, were extracted from EHRs.
Machine learning models incorporating microbiome and clinical data were utilized to predict LC and to identify symptom clusters, providing valuable insights into the heterogeneity of the condition.
Study results The study analyzed 947 stool samples collected from 799 participants, including 380 SARS-CoV-2-positive individuals and 419 negative controls. Of the SARS-CoV-2-positive group, 80 patients developed LC during a one-year follow-up period.
Participants were categorized into three groups for analysis: LC, non-LC (SARS-CoV-2-positive without LC), and SARS-CoV-2-negative. Baseline characteristics revealed significant differences between these groups. LC participants were predominantly female and had more baseline comorbidities compared to non-LC participants.
The SARS-CoV-2-negative group was older, with higher antibiotic use and vaccination rates. These variables were adjusted for in subsequent analyses.
During acute infection, gut microbiome diversity differed significantly between groups. Alpha diversity was lower in SARS-CoV-2-positive participants (LC and non-LC) than in SARS-CoV-2-negative participants.
Beta diversity analyses revealed distinct microbial compositions among the groups, with LC patients exhibiting unique microbiome profiles during acute infection.
Specific bacterial taxa, including Faecalimonas and Blautia, were enriched in LC patients, while other taxa were predominant in non-LC and negative participants. These findings indicate that gut microbiome composition during acute infection is a potential predictor for LC.
Temporal analysis of gut microbiome changes between the acute and post-acute phases revealed significant individual variability but no cohort-level differences, suggesting that temporal changes do not contribute to LC development.
However, machine learning models demonstrated that microbiome data during acute infection, when combined with clinical variables, predicted LC with high accuracy. Microbial predictors, including species from the Lachnospiraceae family, significantly influenced model performance.
Symptom analysis revealed that LC encompasses heterogeneous clinical presentations. Fatigue was the most prevalent symptom, followed by dyspnea and cough.
Cluster analysis identified four LC subphenotypes based on symptom co-occurrence: gastrointestinal and sensory, musculoskeletal and neuropsychiatric, cardiopulmonary, and fatigue-only.
Each cluster exhibited unique microbial associations, with the gastrointestinal and sensory clusters showing the most pronounced microbial alterations. Notably, taxa such as those from Lachnospiraceae and Erysipelotrichaceae families were significantly enriched in this cluster.
Conclusions To summarize, this study demonstrated that SARS-CoV-2-positive individuals who later developed LC exhibited distinct gut microbiome profiles during acute infection. While prior research has linked the gut microbiome to COVID-19 outcomes, few studies have explored its predictive potential for LC, particularly in outpatient cohorts.
Using machine learning models, including artificial neural networks and logistic regression, this study found that microbiome data alone predicted LC more accurately than clinical variables, such as disease severity, sex, and vaccination status.
Key microbial contributors included species from the Lachnospiraceae family, such as Eubacterium and Agathobacter, and Prevotella spp. These findings highlight the gut microbiome’s potential as a diagnostic tool for identifying LC risk, enabling personalized interventions.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference: Preliminary scientific report. Isin Y. Comba, Ruben A. T. Mars, Lu Yang, et al. (2024) Gut Microbiome Signatures During Acute Infection Predict Long COVID, bioRxiv. doi:https://doi.org/10.1101/2024.12.10.626852. www.biorxiv.org/content/10.1101/2024.12.10.626852v1.full
#mask up#public health#wear a mask#pandemic#wear a respirator#covid#still coviding#covid 19#coronavirus#sars cov 2#long covid#AI
36 notes
·
View notes
Text
i'm leafing through another paper for my butterfly research and this has the weirdest organization i've ever seen... why are all of the figures at the end of the pdf?? why aren't they labelled??? and why are the pages they're on at different aspect ratios????? the worst thing is that this paper actually has some cool, relevant stuff so i'll probably have to use it :[
here it is, in case anyone's curious: https://www.biorxiv.org/content/biorxiv/early/2024/02/08/2024.02.07.579046.full.pdf
#melonposting#this research paper is driving me nuts#people need to shut up about nymphalids i swear to god. I DON'T CAAAARE /silly#please just give me a nice pierid. a papillionid even. anything please
5 notes
·
View notes