#Cluster sampling analysis
Explore tagged Tumblr posts
Text
Cluster Sampling: Types, Advantages, Limitations, and Examples
Explore the various types, advantages, limitations, and real-world examples of cluster sampling in our informative blog. Learn how this sampling method can help researchers gather data efficiently and effectively for insightful analysis.
#Cluster sampling#Sampling techniques#Cluster sampling definition#Cluster sampling steps#Types of cluster sampling#Advantages of cluster sampling#Limitations of cluster sampling#Cluster sampling comparison#Cluster sampling examples#Cluster sampling applications#Cluster sampling process#Cluster sampling methodology#Cluster sampling in research#Cluster sampling in surveys#Cluster sampling in statistics#Cluster sampling design#Cluster sampling procedure#Cluster sampling considerations#Cluster sampling analysis#Cluster sampling benefits#Cluster sampling challenges#Cluster sampling vs other methods#Cluster sampling vs stratified sampling#Cluster sampling vs random sampling#Cluster sampling vs systematic sampling#Cluster sampling vs convenience sampling#Cluster sampling vs multistage sampling#Cluster sampling vs quota sampling#Cluster sampling vs snowball sampling#Cluster sampling steps explained
0 notes
Text
Down Under - Part 1
Series masterlist

Part 1
“The epicentre of the outbreak,” Steve was saying to his bleary-eyed team, “appears to be here.” His long pointer made a thwack as it struck the wall map somewhere in the south of Australia. His accompanying PowerPoint ticked over to a slide showing a photo of a single road running through a smattering of houses, deserted save for a large mob of kangaroos. Nice shot, you thought, as you covered a yawn with the back of your hand.
“The village of Hall’s Gap,” Steve continued, “population 496. The Victorian Premier’s Office has been in touch with us tonight – that is, this morning –"
He pulled his wrist away from his face and blinked rapidly in an effort to check his watch. 4.30 am.
“Ahem. What I mean to say is, the local government has asked us to investigate what seems to be an outbreak of an unusual contagious illness. Fortunately, the remoteness of the locale means that the infection is so far contained to this small township. However, speed and discretion remain of the utmost importance.”
Your sleepy mind began to catch on. A contagion outbreak? In Victoria? An Australian mission... You nervously tried to blink away some of the fatigue. You were the obvious choice – a local, an ex-pat. Am I about to be sent home?
Bruce stood, drawing eyes to him in the semi-light afforded by the projector. “Ah – yeah,” he said. “Sorry to get you all up at this hour. But the faster we get in, find the source, and treat the patients, the better chance we have of eradicating it.”
“Hang on,” Barton interrupted, rubbing an eye, “hang on. Just back up a minute. What exactly do you mean, “unusual illness”?”
“Ah – yeah,” Banner said again, his face pink. “I’ll - I’ll just show you.” The PowerPoint ticked over again, and Steve averted his eyes.
This time it was footage. The scene was grainy and captured from above, as though on a cheap security camera; it looked like the front room of a bank. Clusters of bodies, dozens of them, writhed on – or against – every surface. There was no sound, but there was also no mistaking what they were doing.
The conference room was suddenly wide awake.
“Wait...” Natasha spoke to Bruce without moving her eyes from the scene. “Are they?...”
“Involved in coitus, yes,” Steve answered instead, his gaze still resolutely at the floor. “The major symptom of infection is what you see here: an insatiable… desire. For copulation.” He swallowed. “For sexual intercourse.”
Voices broke out across the room. “Brother,” you heard Loki chuckle, “does it not remind you of that party we attended on Vanaheim?”
Rogers spoke over the noise, having overcome his embarrassment. “The repercussions of infection are serious. We mean truly insatiable; patients are forgetting to eat, drink, or sleep. We believe several lives have been lost.”
Muted respect fell over the room, and Bruce spoke again. “I need to get in there and collect a sample in order to prepare treatment options – ideally a vaccine.”
You finally found voice to speak. “Can’t Australia just send you a sample? Why do we need to go in?”
“Great idea,” Tony broke in. “Except that no one who’s entered Hall’s Gap in the past week has come out.”
Steve took over again. “We suspect this is a Hydra bioweapon pilot, possibly released from a hidden location in the nearby national park.” Another slide, this time of picturesque wilderness: mountain streams and gushing waterfalls framed by towering eucalypts and sheer rockfaces. “To that end, our objectives are two-fold. Collect a sample for analysis, and find and neutralise the Hydra base.”
There was a brief silence before Clint spoke again. “Alright, Cap. Who’s going in?”
“It will just be four of you at first; a small group will move faster.” Steve looked directly at you. “As our resident Australian, Agent, I want you on the ground.” You had been expecting the order, but a pit still instantly formed in your stomach. “A local SHIELD operative will meet you and guide you in. Banner will obviously join you… As will our Asgardian brothers here.”
Thor rolled his shoulders back and gave a pompous nod, but Loki narrowed his eyes. “Why?” he asked.
“You never get sick,” Stark cut in again. “Remember that flu that ripped through here in February? The two of you didn’t even sneeze. We don’t know what this is, but you’re the safest bet when it comes to any degree of innate immunity.”
“The rest of us will wait here for your signal,” Rogers continued. “Any sign of Hydra – any suggestion that you might need support – we’ll be on our way.”
“When do we leave?” you asked.
Steve checked his watch again. “An hour.” He squinted. “Make that – forty-six minutes.”

You always hated take-off. The familiar plummet of your stomach as the Quinjet rose into the air; the crushing embrace as it accelerated to full speed. But once you were safely at cruising altitude and could move about again, flying wasn’t so bad. You sat next to Banner as he skillfully piloted the aircraft across the Pacific Ocean, feeling your apprehension grow with every passing mile of open sea.
Focus on the mission, you thought.
“Bruce, if no one has made it back out of Hall’s Gap, how do we know what’s going on there? How did we get that footage?”
“It’s a cloud-recording. When State officials realised no one in town was answering a phone, let alone leaving the area, they accessed the bank’s security footage.” He grinned, but it was mirthless. “Bet they weren’t expecting to see that.”
“So, what – it’s a virus?”
“More likely a fungal pathogen,” Bruce replied. “There’s a cordyceps fungus that does something similar to ghost moths in the Himalayas. I’ve got some generic antifungal meds that we’ll all take as a precaution, but I can’t develop a proper vaccine until I’ve got a sample.”
“How do you get a sample?”
“From infected brain tissue,” Bruce said grimly.
You were interrupted by a deep yelp from behind, and you turned to see Thor shaking out his right hand as though stung. The brothers were passing time with a game that looked like a combination of rock-paper-scissors and bloody knuckles. Loki leaned back in his seat, his cat-got-the-cream expression widening. One long, leather-clad leg stretched out into the aisle; the other bent at the knee so that his foot could rest on the seat in front. You could see the raised outline of his quadriceps. He lifted his arms to settle his hands behind his head, the card of his slender fingers through his own hair making you squirm. Why are the pretty ones always such dickheads?
You mentally shook yourself. Loki’s smarm and sex appeal were irrelevant. People were dying.
Rage flared within you. How dare they. Hydra had targeted Australia not because it posed a threat, or because the location gave them a tactical advantage. It was because a test release of a bioweapon in a place like Hall’s Gap was easy to hide.
Remote. Wild. Dangerous.
You pushed Loki’s long limbs out of your mind. Without your full concentration, the mission could be deadly.

It was after midnight local time when the Quinjet began descending.
“Ten minutes, guys,” Banner said, as the altimeter rapidly ticked down.
Loki stood, making a big show of stretching, his leather armour creaking. He caught you watching him and gave a wolfish grin, then a small shrug of his left shoulder.
You almost jumped in surprise. Green light licked up his body, and the black leather was replaced by sensible, climate-appropriate clothing: a lightweight collared shirt open over a tight, V-necked tank top, and moleskin hiking pants. A small triangle of pale flesh was visible at his shoulder where his layers left a gap. You had to make a conscious effort to close your mouth. Sensible, but hot as hell. I bet those pants make his ass look amazing.
He winked at you from under the wide brim of a dark-brown Akubra, resplendent with what looked like kookaburra feathers, as Banner landed on a grassy flat at the fringe of the Australian desert.
“We’ll sleep here the rest of the night,” he said, as the group descended the Quinjet ramp into the warm, moonless night. “There’s a local guide meeting us in the morning, then it’s a day’s hike into Hall’s Gap. Can’t risk flying any closer and being detected, in case Hydra really is nearby. We’ll stay off the roads for the same reason.”
“When you say, ‘here’…” Loki looked around the rough clearing distastefully.
“Loki, you must learn to tolerate the lesser comforts!” Thor’s jovial voice was louder than ever in the abandoned night. “Remember the time you stole away from the Queen’s retinue at Mimisbrunnen because the baths were too cold…”
You followed the sound of running water to a nearby stream, surrounded by the scent of eucalyptus and tea tree. As water trickled into your canteen, leaves rustled; the movement of some large marsupial, disturbed by your presence. It was unexpectedly comforting.
It’s been too long, you thought, as memory flooded your senses. But then, once upon a time, I didn’t think I’d ever be back again.
There was a slapping sound from the group, and a swear word in a foreign language. “What in Hel?!” Thor spluttered. “These biting insects are the size of small birds!” The hiss of an aerosol can quickly followed, as Banner generously doused him in mosquito repellent. You grinned to yourself.
The Quinjet’s lights shut off, leaving the four of you in darkness. You rolled out your sleeping gear some distance from the others, stripped out of most of your clothing, and lay flat on your back in your sleeping bag. Sleep might be a big ask, you thought, as you gazed upwards. The arm of the Milky Way stretched overhead, like a hug from an old friend.
You’d always secretly thought this hemisphere had the superior night sky. You were mentally cataloguing as many southern constellations as you could remember when Loki appeared out of the night beside you. Is he… topless? It was hard to tell in the dark. Maybe he’s just wearing really tight sleepwear. The thought made you press your thighs together.
“May I?” he asked, polite but vaguely entitled. “Thor, of course, is already snoring loudly enough to disturb Valhalla.” You could indeed hear the deep rumble.
“Ah - sure,” you said, surprised. In general, Loki didn’t speak to you. Or anyone, besides Thor, if you didn’t count barbed quips and snarky commentary on the day-to-day operations of the team. You weren’t even sure he knew your name.
He spread another of SHEILD’s high-tech swags out beside you.
“This is your home, yes?” he asked, as he slid into his bedding.
You let out a deep breath. “Yeah. Well, um, not here here. On the coast.”
“You are lucky to be able to return,” he murmured.
You risked a quick glance at him, struck by the sadness in his voice. “I guess so.” He, too, was gazing up at the night sky. “Do you... miss home?”
“Ceaselessly.”
You felt the silence stretch, disconcerted by his honesty. “Um - can I tell you about our stars?”
“I am very familiar with the Midgardian sky.”
“But the sky here is different. Everything’s upside down, for a start.” You pointed to the constellation of Orion, clearly head-down. “See?”
You heard the slight smile in his voice as he said, “I see. What else?”
Speaking quietly, you pointed out all the familiar sky-marks you had found when you’d first laid down. "It's a pity the SHEILD tactical goggles don’t work very well for the sky – too specialised for detection and warfare, I guess,” you said. A thought occurred to you. “I don’t suppose you’ve got a pair of binoculars in that magic pocket of yours?”
Loki either smiled or grimaced – you could only see the glint of his teeth. “I can do better than that,” he muttered, almost to himself.
With a faint fizzling sound and a flash of silver, the entire night sky blossomed into colour and light.
It was as though you were lying under an enormous telescope dome. Your eyes could discern individual stars of the Omega Centauri cluster, or the spectacular colour and shape of the Carina nebula, or any of a hundred other astronomical wonders suddenly visible to you from horizon to horizon.
You glanced at Loki again. His sky was casting enough light to see him clearly now; he lay with his arm under his head and a serene smile on his face.
“Thank you,” you said softly.
“My pleasure,” he murmured, not taking his eyes off the scene above.
You lay there, the two of you, gazing upwards in silent wonder. You thought about what it meant to be home, why you had left so many years ago… How it might feel to have no home to return to. Until finally, just as Loki’s breathtaking illusion began to fade, you fell asleep.

Part 2
Tags in comments xx
#loki#loki x female reader#loki x reader#loki x you#loki fic#loki smut#avenger loki#sex pollen#(eventually)
238 notes
·
View notes
Text
Also preserved in our archive
By Vijay Kumar Malesu
In a recent pre-print study posted to bioRxiv*, a team of researchers investigated the predictive role of gut microbiome composition during acute Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection in the development of Long Coronavirus Disease (Long COVID) (LC) and its association with clinical variables and symptom clusters.
Background LC affects 10–30% of non-hospitalized individuals infected with SARS-CoV-2, leading to significant morbidity, workforce loss, and an economic impact of $3.7 trillion in the United States (U.S.).
Symptoms span cardiovascular, gastrointestinal, cognitive, and neurological issues, resembling myalgic encephalomyelitis and other post-infectious syndromes. Proposed mechanisms include immune dysregulation, neuroinflammation, viral persistence, and coagulation abnormalities, with emerging evidence implicating the gut microbiome in LC pathogenesis.
Current studies focus on hospitalized patients, limiting generalizability to milder cases. Further research is needed to explore microbiome-driven predictors in outpatient populations, enabling targeted diagnostics and therapies for LC’s heterogeneous and complex presentation.
About the study The study was approved by the Mayo Clinic Institutional Review Board and recruited adults aged 18 years or older who underwent SARS-CoV-2 testing at Mayo Clinic locations in Minnesota, Florida, and Arizona from October 2020 to September 2021. Participants were identified through electronic health record (EHR) reviews filtered by SARS-CoV-2 testing schedules.
Eligible individuals were contacted via email, and informed consent was obtained. Of the 1,061 participants initially recruited, 242 were excluded due to incomplete data, failed sequencing, or other issues. The final cohort included 799 participants (380 SARS-CoV-2-positive and 419 SARS-CoV-2-negative), providing 947 stool samples.
Stool samples were collected at two-time points: weeks 0–2 and weeks 3–5 after testing. Samples were shipped in frozen gel packs via overnight courier and stored at −80°C for downstream analyses. Microbial deoxyribonucleic acid (DNA) was extracted using Qiagen kits, and metagenomic sequencing was performed targeting 8 million reads per sample.
Taxonomic profiling was conducted using Kraken2, and functional profiling was performed using the Human Microbiome Project Unified Metabolic Analysis Network (HUMAnN3).
Stool calprotectin levels were measured using enzyme-linked immunosorbent assay (ELISA), and SARS-CoV-2 ribonucleic acid (RNA) was detected using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Clinical data, including demographics, comorbidities, medications, and symptom persistence, were extracted from EHRs.
Machine learning models incorporating microbiome and clinical data were utilized to predict LC and to identify symptom clusters, providing valuable insights into the heterogeneity of the condition.
Study results The study analyzed 947 stool samples collected from 799 participants, including 380 SARS-CoV-2-positive individuals and 419 negative controls. Of the SARS-CoV-2-positive group, 80 patients developed LC during a one-year follow-up period.
Participants were categorized into three groups for analysis: LC, non-LC (SARS-CoV-2-positive without LC), and SARS-CoV-2-negative. Baseline characteristics revealed significant differences between these groups. LC participants were predominantly female and had more baseline comorbidities compared to non-LC participants.
The SARS-CoV-2-negative group was older, with higher antibiotic use and vaccination rates. These variables were adjusted for in subsequent analyses.
During acute infection, gut microbiome diversity differed significantly between groups. Alpha diversity was lower in SARS-CoV-2-positive participants (LC and non-LC) than in SARS-CoV-2-negative participants.
Beta diversity analyses revealed distinct microbial compositions among the groups, with LC patients exhibiting unique microbiome profiles during acute infection.
Specific bacterial taxa, including Faecalimonas and Blautia, were enriched in LC patients, while other taxa were predominant in non-LC and negative participants. These findings indicate that gut microbiome composition during acute infection is a potential predictor for LC.
Temporal analysis of gut microbiome changes between the acute and post-acute phases revealed significant individual variability but no cohort-level differences, suggesting that temporal changes do not contribute to LC development.
However, machine learning models demonstrated that microbiome data during acute infection, when combined with clinical variables, predicted LC with high accuracy. Microbial predictors, including species from the Lachnospiraceae family, significantly influenced model performance.
Symptom analysis revealed that LC encompasses heterogeneous clinical presentations. Fatigue was the most prevalent symptom, followed by dyspnea and cough.
Cluster analysis identified four LC subphenotypes based on symptom co-occurrence: gastrointestinal and sensory, musculoskeletal and neuropsychiatric, cardiopulmonary, and fatigue-only.
Each cluster exhibited unique microbial associations, with the gastrointestinal and sensory clusters showing the most pronounced microbial alterations. Notably, taxa such as those from Lachnospiraceae and Erysipelotrichaceae families were significantly enriched in this cluster.
Conclusions To summarize, this study demonstrated that SARS-CoV-2-positive individuals who later developed LC exhibited distinct gut microbiome profiles during acute infection. While prior research has linked the gut microbiome to COVID-19 outcomes, few studies have explored its predictive potential for LC, particularly in outpatient cohorts.
Using machine learning models, including artificial neural networks and logistic regression, this study found that microbiome data alone predicted LC more accurately than clinical variables, such as disease severity, sex, and vaccination status.
Key microbial contributors included species from the Lachnospiraceae family, such as Eubacterium and Agathobacter, and Prevotella spp. These findings highlight the gut microbiome’s potential as a diagnostic tool for identifying LC risk, enabling personalized interventions.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference: Preliminary scientific report. Isin Y. Comba, Ruben A. T. Mars, Lu Yang, et al. (2024) Gut Microbiome Signatures During Acute Infection Predict Long COVID, bioRxiv. doi:https://doi.org/10.1101/2024.12.10.626852. www.biorxiv.org/content/10.1101/2024.12.10.626852v1.full
#mask up#public health#wear a mask#pandemic#wear a respirator#covid#still coviding#covid 19#coronavirus#sars cov 2#long covid#AI
36 notes
·
View notes
Text
"In the most comprehensive national study since the onset of the COVID-19 pandemic, a team of researchers that includes a Rutgers-organized consortium of pediatric sites has concluded that long COVID symptoms in children are tangible, pervasive, wide ranging and clinically distinct within specific age groups. Results of the study, funded by the National Institutes of Health (NIH), are published in the Journal of the American Medical Association. “We have convincing evidence that COVID-19 is not just a mild, benign illness for children,” said Lawrence C. Kleinman, a professor of pediatrics and population health expert at Rutgers Robert Wood Johnson Medical School (RWJMS) and the study’s third co-author. “There are children who are clearly disabled by long COVID for long periods of time.” In the early stages of the pandemic, a myth arose and was perpetuated suggesting that because children often developed only mild cases of COVID-19, the risks for young patients were not serious. But this conjecture dissipated amid a rash of data demonstrating that a few children infected with COVID-19 will get very sick and others will suffer an array of health complications long after initial exposure. Broadly defined, long COVID includes symptoms, signs, and conditions – such as aches, fatigue, memory loss and stomach pain – that develop, persist or relapse more than a month after a COVID-19 infection. Worldwide, an estimated 65 million people, including children, live with long COVID. Until recently, most research into COVID-19’s lingering effects focused on adults. To quantify long COVID’s impact on children and determine whether symptoms experienced by the youngest COVID-19 patients differ by age group and from adults, Kleinman and more than 140 researchers throughout the United States crunched data from NIH’s Researching COVID to Enhance Recovery (RECOVER) Initiative, a national effort to survey COVID-19’s long-term impact. As part of RECOVER, caregivers for 5,367 children (898 school-aged children and 4,469 adolescents) completed online surveys about their children’s health in time for this data analysis. Roughly 86 percent of the sample had previously been infected with COVID-19, while 14 percent – the control group – had not. The survey assessed 74 known and potential long COVID-19 symptoms across nine domains: eyes, ears, nose and throat; heart and lungs; gastrointestinal; dermatologic; musculoskeletal; neurologic; behavioral and psychological; menstrual; and general. By analyzing the responses, researchers found 45 percent of COVID-19 infected school-age children (ages 6 to 11) reported at least one prolonged symptom after initial recovery versus 33 percent of uninfected children. Thirty-nine percent of COVID-19 infected adolescents (ages 12 to 17) reported one prolonged symptom, compared with 27 percent of uninfected adolescents. These differences implicate the virus as a likely causal factor, rather than just having lived through the pandemic. Long COVID symptoms in children also were clustered in patterns distinct from adults and from each other. For instance, the most common symptom in adolescents was loss of smell and taste, followed by low energy, muscle aches and fatigue. For school-age children, memory and focus issues topped the list, followed by stomach pain, headaches and back or neck pain. Children experienced prolonged symptoms after COVID-19 infection “in almost every organ system, with the vast majority having multisystem involvement,” the authors wrote."
oh hey, remember a couple of years ago during the omicron wave when multisystem inflammatory syndrome was driving a lot of kids to go to the hospital?
29 notes
·
View notes
Text

The first 3D view of the formation and evolution of globular clusters
A study published today in Astronomy & Astrophysics marks a significant milestone in our understanding of the formation and dynamical evolution of multiple stellar populations in globular clusters (spherical and very compact stellar agglomerates typically populated by 1–2 million stars). This pioneering study, conducted by a group of researchers from the National Institute for Astrophysics (INAF), the University of Bologna, and Indiana University, is the first to perform a 3D kinematic analysis of multiple stellar populations for a representative sample of 16 globular clusters in our Galaxy. It provides a groundbreaking observational description of their kinematic properties (i.e., how stars move within globular clusters) and their long-term evolution from the formation to the present day.
Emanuele Dalessandro, researcher at INAF in Bologna, lead author of the article and coordinator of the working group, explains: "Understanding the physical processes behind the formation and early evolution of globular clusters is one of the most fascinating and debated astrophysical questions of the past 20–25 years. The results of our study provide the first solid evidence that globular clusters formed through multiple star formation events and place fundamental constraints on the dynamical path followed by the clusters throughout their evolution. These results were made possible by a multi-diagnostic approach and the combination of state-of-the-art observations and dynamic simulations."
The study highlights that the kinematic differences between multiple populations are key to understanding the formation and evolution mechanisms of these ancient structures.
With ages that can reach 12-13 billion years (thus dating back to the dawn of the cosmos), globular clusters are among the first systems to form in the Universe. They represent a typical population of all galaxies. They are compact systems (with masses of several hundred thousand solar masses and sizes of a few parsecs), and they can be observed even in distant galaxies.
"Their astrophysical significance is huge," says Dalessandro, "because they not only help us to test cosmological models of the formation of the Universe due to their age but also provide natural laboratories for studying the formation, evolution, and chemical enrichment of galaxies." Despite globular clusters have been studied for over a century, recent observational results show that our knowledge is still largely incomplete.
"Results obtained in the last two decades have unexpectedly shown that globular clusters consist of more than one stellar population: a primordial one, with chemical properties similar to other stars in the Galaxy, and another with anomalous chemical abundances of light elements such as helium, oxygen, sodium, and nitrogen," says Mario Cadelano, researcher at the Department of Physics and Astronomy at the University of Bologna and INAF associate, one of the authors of the study. "Despite the large number of observations and theoretical models aimed at characterising these populations, the mechanisms regulating their formation are still not understood."
The study is based on the measurement of 3D velocities, i.e., the combination of proper motions and radial velocities, obtained with the ESA Gaia telescope and with data from, among others, the ESO VLT telescope, primarily as part of the MIKiS survey (Multi Instrument Kinematic Survey), a spectroscopic survey specifically aimed at exploring the internal kinematics of globular clusters. The use of these telescopes, from space and the ground, has provided an unprecedented 3D view of the velocity distribution of stars in the selected globular clusters.
The analysis reveals that stars with different abundances of light elements are characterised by different kinematic properties, such as rotational velocities and orbital distributions.
"In this work, we analysed in detail the motion of thousands of stars within each cluster," adds Alessandro Della Croce, a PhD student at INAF in Bologna. "It quickly became clear that stars belonging to different populations have distinct kinematic properties: stars with anomalous chemical composition tend to rotate faster than the others within the cluster and progressively spread from the central regions to the outer ones."
The intensity of these kinematic differences depends on the dynamical age of globular clusters. "These results are consistent with the long-term dynamical evolution of stellar systems, in which stars with anomalous chemical abundances form more centrally concentrated and rotate more rapidly than the standard ones. This, in turn, suggests that globular clusters formed through multiple star formation episodes and provides an important piece of information in defining the physical processes and timescales underlying the formation and evolution of massive stellar clusters," Dalessandro emphasises.
This new 3D view of the motion of stars within globular clusters provides an unprecedented and fascinating framework for the formation and dynamical evolution of these intriguing systems. It also helps to clarify some of the most complex mysteries surrounding the origin of these ancient structures.
IMAGE: Image gallery of the 16 globular clusters analysed in order of difference in the kinematic properties observed between the multiple stellar populations. Credit Credits: ESA/Hubble - ESO - SDSS
11 notes
·
View notes
Text
Another New Disease That Trump is Weakening US Response To
We remember how COVID-19 was allowed to run rampant in the United States for 51 days before Trump bothered to declare a state of emergency on Friday the 13th of March in 2020.
With newly installed incompetent MAGA administrators and deep cuts to public health, the US is now even less prepared for any new infectious disease to hit the country.
A deadly disease which doesn't even have a name yet has hit Équateur province in the Democratic Republic of the Congo (DRC). The World Health Organization mentioned it prominently in its 16 February 2025 Bulletin.
Unknown Disease in the Democratic Republic of the Congo: The Democratic Republic of the Congo is facing multiple public health and humanitarian crises. In its northwestern Équateur Province, two clusters of cases and deaths from an unknown disease have emerged, resulting in hundreds of cases and dozens of deaths. The outbreak, which has seen cases rise rapidly within days, poses a significant public health threat. The exact cause remains unknown, with Ebola and Marburg already ruled out, raising concerns about a severe infectious or toxic agent. Key challenges include the rapid progression of the disease, with nearly half of the deaths occurring within 48 hours of symptom onset in one of the affected health zones, and an exceptionally high case fatality rate in another. Urgent action is needed to accelerate laboratory investigations, improve case management and isolation capacities, and strengthen surveillance and risk communication. [ ... ] Specimens from thirteen cases, including 12 blood samples from active cases and one swab from a deceased individual, were collected and sent to the National Institute of Biomedical Research (INRB) in Kinshasa for analysis on 11 February 2025. Test results released on 13 February 2025, showed that all samples were negative for Ebola and Marburg viruses by polymerase chain reaction (PCR). Differential diagnosis under investigation include malaria, viral haemorrhagic fever, food or water poisoning, typhoid fever, and meningitis.
So essentially, Ebola and Marburg virus have been ruled out. We do know that the fatality rate is way high.
The situation in Équateur Province presents significant public health risk, with two clusters of an unknown disease causing high morbidity and mortality. The overall case fatality ratio (12.2%), particularly high in Bolomba Health Zone (66.7%), and the rapid disease progression raise concerns about a severe infectious or toxic agent.
We should point out that this is in the northwest of the DRC – not the east where there is a de facto war going on.

The Obama administration had great success combating Ebola. There were just 11 fatalities in the US with just 2 being from infections contracted inside the country during the 2013-2014 outbreak.
Obama's national security team put together a pandemic playbook based on their success to be used by succeeding administrations. Of course Donald Trump ignored it and the result was hundreds of thousands of additional American deaths from COVID-19.
Obama team left pandemic playbook for Trump administration, officials confirm
If this new unnamed disease spreads to the US, don't expect Trump to respond competently to it.
#new disease#democratic republic of the congo#drc#équateur#world health organization#pandemic#obama administration#pandemic playbook#donald trump#trump's incompetent response to covid-19#public health#infection
13 notes
·
View notes
Text
What Is PluriCell? Everything You Need to Know About This All-in-One Cell Culture and Filtration System
In laboratories that handle tissue digestion, live-cell assays, and organism tracking, combining cell separation and cell culture steps can save time and reduce contamination risks. That’s where PluriCell, a multi-well Lab Cell Strainer system, offers a game-changing solution. It allows filtration and culturing within the same plate setup—minimizing handling, maximizing throughput, and supporting high-precision assays.
Whether you're working with nematodes, single cells, or small multicellular organisms, PluriCell simplifies the workflow by integrating size-based filtration directly into multi-well plates. For labs looking to improve efficiency without compromising on accuracy, understanding how PluriCell works—and why it’s different from standard cell strainers—is the first step to adopting a better solution.
What Is PluriCell? A Quick Overview
PluriCell is a sample preparation and assay tool designed for researchers working with single-cell and multi-cellular organisms. The system includes nylon mesh-based strainers pre-fitted into standard well plate formats (6-, 12-, 24-, and 48-well). Each strainer plate fits snugly into corresponding well plates, allowing size-based cell separation and in-well culturing to occur simultaneously.
The mesh comes in multiple sizes—ranging from 5 µm to 500 µm—offering flexibility for diverse cell types or organisms. From isolating parasites to preparing single-cell suspensions for screening, PluriCell covers a wide range of applications while maintaining a compact footprint and simple setup.
Most importantly, PluriCell is not just a cell strainer—it’s a complete in-well assay platform that supports live tracking, treatment, and endpoint measurement of cultured samples.
How PluriCell Combines Filtration and Culture
Traditional workflows require multiple steps for size separation and cell plating. Samples are often strained through mesh filters, transferred into culture plates, and then moved again for drug testing or analysis. These transfers increase the risk of losing viable cells, introduce inconsistencies, and lengthen protocol times.
PluriCell changes that. It enables the particle separation process to occur directly in the same environment where the culture or assay will take place.
Key Benefits:
One step to filter and culture: No more transferring between mesh filters and plates.
Reduced sample loss: Cells or organisms are never handled more than necessary.
Higher reproducibility: Less manual handling leads to more consistent results.
Supports live assays: Migration, paralysis, and viability studies can be done without removing the sample.
Plate Format and Mesh Options
The PluriCell system is compatible with common lab equipment and comes in a range of configurations:
Multi-Well Plate Formats:
6-well
12-well
24-well
48-well
These options allow labs to scale up or down based on throughput needs, from small pilot studies to large screening assays.
Mesh Sizes Available:
Fine Meshes: 5, 10, 20, and 40 µm for single-cell filtration and smaller organisms.
Intermediate Meshes: 70 and 100 µm, ideal for cell clusters and small larvae.
Coarse Meshes: 200 and 500 µm for nematodes and larger multicellular samples.
This mesh variety allows labs to select exactly the right tool for their target sample. Instead of compromising by using an oversized or undersized filter, labs can match mesh size to assay requirements, enabling more accurate particle separation techniques.
Cascade Straining in a Multi-Well Format
One standout feature of PluriCell is how easily it enables cascade straining. Cascade straining refers to passing samples through a series of filters with decreasing pore sizes. This technique is especially helpful when dealing with tissues or samples containing a mix of particles, cell types, or organisms.
With PluriCell’s multi-well setup, each well can hold a strainer of a different mesh size. Researchers can load the sample once and allow gravity or light pipetting to filter the sample through wells in a defined sequence.
Example Workflow:
Start with a 500 µm mesh to remove large debris.
Next, use a 100 µm mesh for small tissue clusters or larger organisms.
Finish with a 40 µm mesh to isolate single cells or small larvae.
This setup provides fast and easy cell separation without repeated sample handling or multiple filter tools. More importantly, the entire cascade can happen within a single plate—saving time and preserving sterility.
Applications in Research and Screening
PluriCell isn’t just about convenience. It also enables entirely new types of experiments. Because cells or organisms remain in the well after filtration, they can be directly exposed to drugs, dyes, or environmental changes.
Common Use Cases:
Parasite migration assays
Viability and toxicity testing
Stem cell and primary cell culture
High-throughput screening (HTS)
Live imaging of multicellular organisms
Labs that handle fragile specimens like C. elegans, planarians, or patient-derived cell types benefit most. The system minimizes stress on the sample while maintaining clarity and access for measurement tools.
Why Mesh Integrity and Build Quality Matter
For labs used to standard lab cell strainers, PluriCell may appear similar—but its mesh quality sets it apart.
Uniform nylon mesh ensures consistent flow rates and pore sizing.
No deformation or mesh slippage, even after repeated handling.
Sterile and single-use to maintain culture conditions and reduce contamination risk.
PluriCell vs. Traditional Culture + Filtration Setups
When comparing PluriCell to standard lab filtration and culture workflows, several distinct advantages stand out:
Filtration Method Traditional workflows rely on external tools like syringe filters or cap strainers for filtration, which must then be manually transferred to culture plates. PluriCell integrates filtration directly into the well plate, eliminating the need for separate filtering devices.
Sample Handling and Transfers Conventional methods involve multiple handling steps, increasing time and risk of error. With PluriCell, filtration and culture occur in the same well, streamlining the process into a single step.
Contamination Risk Each manual transfer in traditional workflows increases the likelihood of introducing contaminants. PluriCell reduces this risk with its sealed design and sterile, ready-to-use format.
Assay Readiness In standard setups, separate tools are required to move from filtration to culturing or assays. PluriCell is fully integrated—after filtration, samples remain in place for immediate treatment, observation, or analysis.
Scalability and Throughput Traditional methods require manual adjustments and more consumables to scale up. PluriCell supports high-throughput formats with 6-, 12-, 24-, and 48-well options, making it easy to expand workflows without adding complexity.
How PluriCell Saves Time in Lab Workflows
Time is a valuable resource in any lab. The fewer the steps, the less room there is for error, delays, or sample loss. PluriCell’s design reflects an understanding of this.
Here’s how it streamlines your process:
Quick setup: No assembling mesh tools or transferring between formats.
Fewer consumables: Plates, filters, and supports are combined in one product.
Immediate use after filtration: Cells or organisms are already in place for assays or culture.
This speed and simplicity are what make PluriCell a practical upgrade from standalone lab cell strainer tools.
Conclusion
PluriCell offers more than filtration—it transforms how researchers approach cell preparation and culture. By combining mesh-based separation with multi-well assay formats, it simplifies complex protocols and supports more reliable data generation.
Its compatibility with various mesh sizes, plate formats, and cell types makes it a versatile tool for modern labs. Whether you're filtering single cells, tracking nematode migration, or running live-cell drug response assays, PluriCell delivers consistent, clean, and assay-ready results.
For labs serious about improving throughput and reducing variability, Lab Cell Strainer like PluriCell represent a powerful upgrade over traditional tools. Explore the full range of PluriCell systems today to streamline your workflow and make every assay count.
#particle filtration#cascade straining#cell enrichment techniques#syringe strainer#lab cell strainer
2 notes
·
View notes
Note
Can I ask for 17. noticing their individual quirks from the blossoming romance prompt list with Simpatico? <3
WHAT A COINCIDENCE THAT I STARTED REWRITING LIKE 3 HOURS BEFORE I SAY THE POST ABOUT YOUR BIRTHDAY!! Anyways, Happy Belated Birthday!!! Enjoy some simpatico nonsense:)
Ao3 Link Here
Perceptor narrowed his optics down at the pile of clutter before him. Clutter was a kind, professional, polite way of describing explosive havoc of disorder and chaos that made up the dimensions of Brainstorm’s desk. Disgusting was another word that came to mind.
::How do you live like this?::
::Oh please, let’s not exaggerate. It’s not that bad.:: came Brainstorm’s groan.
It was not an exaggeration. If anything, it was an understatement.
Perceptor’s internal processors had a difficult time distinguishing just what exactly he was looking at. The only way to actually piece through what was on the desk was to deconstruct it layer by layer. A cross-section analysis.
The bottom-most layer- the foundation, if you will -were dried dribbles of fuel intermingled with a noxious dusting of sentiment and dirt. One of Perceptor’s background scanners identified a cluster of granulated particles to be aged candied energon treat crumbs. An entire rust strick made the foundation brick, its sticky residue gluing it to the hard surface of the desk. Perceptor idly pondered if its removal would cause the entire system to fall apart. And while his internal protocols desperately would like the area cleaned, organized and sanitary, he was not willing to find out if his hypothesis was correct.
Cemented to this foundational core layer was the secondary mantle layer. This, from what Perceptor could read, was a scattering of notes all in Brainstorm’s sloppy, near illegible scribble. Tattered napkin bits from Swerve’s and printed notices from Ultra Magnus acted as the canvas for dynamic invention designs, schematics and impossible (and implausible) equations with attached nonsensical theorems. Several datapads acted as structural weights. When flicked on, Perceptor wasn’t sure if he felt amusement, exasperation or a sickly, prickling bashfulness in seeing several of his academic research papers and studies riddled with extensive notes, doodles and elaborations from Brainstorm.
It didn’t take away from the utter disaster that was Brainstorm’s work space but it did soften the blow. Still, Perceptor would prefer if his research wasn’t adding to the disgusting catastrophe that made up Brainstorm’s desk. Perhaps a bookshelf or three would greatly benefit organization.
Level three- the crust -was as troublesome as the other two layers of clutter, if not more prone to disaster by their fragile and incongruous shapes. Trinkets , Brainstorm affectionately called them. Garbage , Perceptor was more keen on describing. In truth, they probably served best as paperweights, however haphazardly placed they were.
The sentimentality was not missed on Perceptor and a part of him could even find the collection charming. Endearing.
Perceptor had bared witness to the slow accumulation over the course of the Lost Light’s journey but had never really taken the time to truly examine them. Now he did, his optics scanning over the seemingly random series of objects: little samples of rock, crystal, fossil collected on pit-stop planets, a Rodi-Star for Temporal Excellence half hanging off the desk, a cluster of thumb drive stocked with films, music, and other media either gifted or stolen from Rewind- Perceptor was still not sure. Little gadgets and doodles from Nautica were in abundance and horrible tiny contractions built by Whirl intermingled with them. There was even a small toy-like bauble on the corner of his desk from Chromedome, Perceptor had been present when the Mnemosurgeon had left it there and Brainstorm never moved it, simply fiddled with it absentmindedly while mulling over his work before throwing it back to the corner of his desk.
All these items, papers and dirt and yet Perceptor still did not actually find what he was looking for.
With a heavy sign, Perceptor responded to the insisting ping in his comms.
::How do you expect me to find anything on your desk?::
Brainstorm’s response was bitingly quick. ::What are you talking about? Everything is organized!::
::It’s garbage, Brainstorm.::
::Use that brilliant mind of yours and you’ll see everything has a purpose.::
::What purpose do Ultra Magnus’s cease orders from 28 cycles ago have?:: Perceptor didn’t dare touch the fragile, lopsided stack in fear of it tumbling down and only adding to the mess.
::They are counterbalances. Don’t move them or the desk will collapse.:: Perceptor had no doubt in the truth of that statement even if its intent was a joke.
::We are cleaning this when you return to the ship.::
::It doesn’t need any cleaning! I know where everything is!:
Perceptor let out a derisive snort. He could picture perfectly the little fluttering of Brainstorm’s ailerons, his hands moving in frustration.
::The tell me where your cathetometer is.::
It was the reason for this call in the first place. For rare occasion, Perceptor had the lab to himself with Brainstorm accompanying Rodimus’s small expedition team. It’s not Perceptor’s fault his colleague forgot his equipment but he was not about to be a complete aft in not assisting. He just wasn’t going to personally dig through Brainstorm’s garbage heap of a desk alone.
::Hmm, if you don’t see it in top it’s probably in one of the drawers.::
Perceptor rounded the desk to see six drawers lining the sides of the desk with three on each side.
::Which one?::
::The left side. I keep the important stuff there.::
Perceptor raised an optic ridge and couldn’t help but ask ::And what do you keep on the right?::
::Come on Percy, let me have a little mystery, a touch in intrigue.::
::Nevermind, I don’t want to know.::
Perceptor didn’t need to be present to know Brainstorm was pouting, blast mask intact or not. Even hundreds of meters between them and Perceptor knew a pouting, sulking Brainstorm anywhere.
::You’re no fun.::
::Yes I am.:: Perceptor replied back as he started with the top drawer, pulling it open only to find it crammed to the brim with even more data pads. All of them pressed together to a block so not even a tiny piece of dust could enter. Perceptor slammed the drawer shut. ::How do you live like this?:: he found himself reiterating.
::Oh, not fun loving Perceptor still complaining about my desk. Is that fun? Cleaning and organizing?::
::You’re a scientist. How do you find anything in this?::
::Tell me how you are fun in explicit detail and I’ll tell you my organizational strategies. We can make a date of it.::
Perceptor snorted as he opened the second drawer. This was filled with several instruments and after some careful digging, he found the cathetometer . ::We can clean your desk together.::
::You must be a hit at the club, Percy. Really. Absolute stud. What moves do you have? The pencil sharpener? The label maker? The file organizer? Actually, you can’t claim that one. Minimus invented and perfected that one. ::
Perceptor could have told Brainstorm at any moment that he had found what the other mech was looking for but, he held onto the tool for a moment, softly smiling to himself as Brainstorm rambled insults to him. It shouldn’t be charming, it shouldn’t be amusing, it shouldn’t bubble up any sort of affection. And yet.
::I’ve seen you dance, Brainstorm. I wouldn’t speak so confidentially with what you’ve demonstrated.::
::Are you saying Minimus is a better dancer than me? Because you surely can be saying that you are a better dancer. I mean, I think you’ll fall apart if you stepped foot on the dance floor.::
::It hasn’t happened yet.::
::When have you been dancing at Swerve’s? Before or after you deep clean and detail your desk every day?::
::Funny.::
Without even thinking about it, Perceptor opened the third drawer. He stopped as it slid open, its few contents rocking in the sway. Recognition lit his processor in a warm, shy heat.
::I’m hilarious. So funny and smart and amazing and talented and resourceful. Speaking of resourceful…did you find the my cathetometer yet? I put googly eyes on it. For personality. Can’t miss it.::
Perceptor felt the warmth spread across his faceplates. ::I did.::
::Oh Percy, I could kiss you. Tailgate is almost back at the ship if you can give it to him. The mods to his hoverboard make him almost as fast as Rodimus. He’s pissed. Anyways I told you it would be easy to find. All my important stuff is.::
Perceptor barely heard a word of what Brainstorm said. Only sending back a short affirmative as he stared at the drawer.
::Perceptor? You alright?::
With a sharp invent, Perceptor closed the drawer shut firmly.
::Perfectly fine. I’ll be ready to pass it off to Tailgate. I’m clearing your schedule for the next cycle. We are cleaning your desk. I can’t work knowing you are working like this.::
::Percy! It’s fine. I don’t need your shitty excuse for a date-::
::It’s not a date.:: Perceptor swiftly cut off. ::This is a work hazard that is being remedied immediately.::
Brainstorm’s response was muted, delayed. ::Okay, okay. We’ll clean it up. I’m sure you’ll have a checklist and everything.::
Perceptor let a small smile come to his face even though he could hear the telltale rumbling of Tailgate’s juiced up hoverboard. ::Of course. You shouldn’t expect anything less from me. If you manage to get it done by shift’s end, I’ll buy you a drink. Maybe if you are lucky, we can dance.::
Brainstorm’s next several responses were streams of incoherent stutters that formed a very excitable agreement. Perceptor didn’t feel the need to continue the chatter as he passed over the tool to Tailgate who only gave him a slightly confused look at his smile. Perceptor didn’t care, not when he knew what lay at the bottom of Brainstorm’s important drawer.
Sentimental fool.
20 notes
·
View notes
Text
Retrospective epidemiologic and genomic surveillance of arboviruses in 2023 in Brazil reveals high co-circulation of chikungunya and dengue viruses
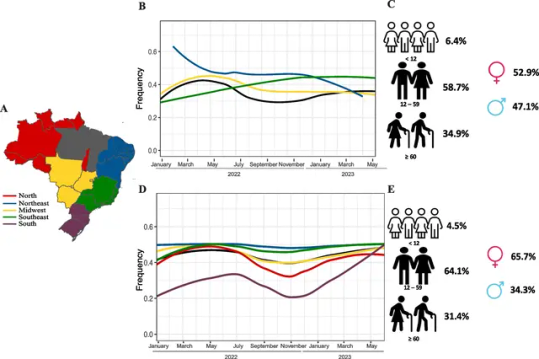
Background
The rapid spread and increase of chikungunya (CHIKV) and dengue (DENV) cases in Brazilian regions in 2023 has raised concerns about the impact of arboviruses on public health. Epidemiological and genomic surveillance was performed to estimate the introduction and spread of CHIKV and DENV in Brazil.
Methods
This study obtained results from the Hermes Pardini (HP), a private medical laboratory, and the Health Department of Minas Gerais state (SES-MG). We investigated the positivity rates of CHIKV and DENV by analyzing the results of 139,457 samples tested for CHIKV (44,029 in 2022 and 95,428 in 2023) and 491,528 samples tested for DENV (163,674 in 2022 and 327,854 in 2023) across the five representative geographical regions of Brazil. Genome sequencing was performed on 80 CHIKV and 153 DENV samples that had been positive for RT-PCR tests.
Results
In our sampling, the data from CHIKV tests indicated that the Northeast region had the highest regional positivity rate in 2022 (58.1%). However, in 2023, the Southeast region recorded the highest positivity rate (40.5%). With regard to DENV, the South region exhibited the highest regional positivity rate in both 2022 (40.8%) and 2023 (22.7%), followed by the Southeast region in both years (34.8% in 2022; 21.4% in 2023). During the first 30 epidemiological weeks of 2023 in the state of Minas Gerais (MG), there was a 5.8-fold increase in CHIKV cases and a 3.5-fold increase in DENV compared to the same period in 2022. Analysis of 151 new DENV-1 and 80 CHIKV genomes revealed the presence of three main clusters of CHIKV and circulation of several DENV lineages in MG. All CHIKV clades are closely related to genomes from previous Brazilian outbreaks in the Northeast, suggesting importation events from this region to MG. We detected the RNA of both viruses in approximately 12.75% of the confirmed positive cases, suggesting an increase of co-infection with DENV and CHIKV during the period of analysis.
Conclusions
These high rates of re-emergence and co-infection with both arboviruses provide useful data for implementing control measures of Aedes vectors and the urgent implementation of public health politics to reduce the numbers of CHIKV and DENV cases in the country.
Read the paper.
#brazil#brazilian politics#politics#science#biology#epidemiology#dengue#chikungunya#image description in alt#mod nise da silveira
2 notes
·
View notes
Text
Inhibition of EIF4E Downregulates VEGFA and CCND1 Expression to Suppress Ovarian Cancer Tumor Progression by Jing Wang in Journal of Clinical Case Reports Medical Images and Health Sciences
Abstract
This study investigates the role of EIF4E in ovarian cancer and its influence on the expression of VEGFA and CCND1. Differential expression analysis of VEGFA, CCND1, and EIF4E was conducted using SKOV3 cells in ovarian cancer patients and controls. Correlations between EIF4E and VEGFA/CCND1 were assessed, and three-dimensional cell culture experiments were performed. Comparisons of EIF4E, VEGFA, and CCND1 mRNA and protein expression between the EIF4E inhibitor 4EGI-1-treated group and controls were carried out through RT-PCR and Western blot. Our findings demonstrate elevated expression of EIF4E, VEGFA, and CCND1 in ovarian cancer patients, with positive correlations. The inhibition of EIF4E by 4EGI-1 led to decreased SKOV3 cell clustering and reduced mRNA and protein levels of VEGFA and CCND1. These results suggest that EIF4E plays a crucial role in ovarian cancer and its inhibition may modulate VEGFA and CCND1 expression, underscoring EIF4E as a potential therapeutic target for ovarian cancer treatment.
Keywords: Ovarian cancer; Eukaryotic translation initiation factor 4E; Vascular endothelial growth factor A; Cyclin D1
Introduction
Ovarian cancer ranks high among gynecological malignancies in terms of mortality, necessitating innovative therapeutic strategies [1]. Vascular endothelial growth factor (VEGF) plays a pivotal role in angiogenesis, influencing endothelial cell proliferation, migration, vascular permeability, and apoptosis regulation [2, 3]. While anti-VEGF therapies are prominent in malignancy treatment [4], the significance of cyclin D1 (CCND1) amplification in cancers, including ovarian, cannot be overlooked, as it disrupts the cell cycle, fostering tumorigenesis [5, 6]. Eukaryotic translation initiation factor 4E (EIF4E), central to translation initiation, correlates with poor prognoses in various cancers due to its dysregulated expression and activation, particularly in driving translation of growth-promoting genes like VEGF [7, 8]. Remarkably, elevated EIF4E protein levels have been observed in ovarian cancer tissue, suggesting a potential role in enhancing CCND1 translation, thereby facilitating cell cycle progression and proliferation [9]. Hence, a novel conjecture emerges: by modulating EIF4E expression, a dual impact on VEGF and CCND1 expression might be achieved. This approach introduces an innovative perspective to impede the onset and progression of ovarian cancer, distinct from existing literature, and potentially offering a unique therapeutic avenue.
Materials and Methods
Cell Culture
Human ovarian serous carcinoma cell line SKOV3 (obtained from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) was cultured in DMEM medium containing 10% fetal bovine serum. Cells were maintained at 37°C with 5% CO2 in a cell culture incubator and subcultured every 2-3 days.
Three-Dimensional Spheroid Culture
SKOV3 cells were prepared as single-cell suspensions and adjusted to a concentration of 5×10^5 cells/mL. A volume of 0.5 mL of single-cell suspension was added to Corning Ultra-Low Attachment 24-well microplates and cultured at 37°C with 5% CO2 for 24 hours. Subsequently, 0.5 mL of culture medium or 0.5 mL of EIF4E inhibitor 4EGI-1 (Selleck, 40 μM) was added. After 48 hours, images were captured randomly from five different fields—upper, lower, left, right, and center—using an inverted phase-contrast microscope. The experiment was repeated three times.
GEPIA Online Analysis
The GEPIA online analysis tool (http://gepia.cancer-pku.cn/index.html) was utilized to assess the expression of VEGFA, CCND1, and EIF4E in ovarian cancer tumor samples from TCGA and normal samples from GTEx. Additionally, Pearson correlation coefficient analysis was employed to determine the correlation between VEGF and CCND1 with EIF4E.
RT-PCR
RT-PCR was employed to assess the mRNA expression levels of EIF4E, VEGF, and CCND1 in treatment and control group samples. Total RNA was extracted using the RNA extraction kit from Vazyme, followed by reverse transcription to obtain cDNA using their reverse transcription kit. Amplification was carried out using SYBR qPCR Master Mix as per the recommended conditions from Vazyme. GAPDH was used as an internal reference, and the primer sequences for PCR are shown in Table 1.
Amplification was carried out under the following conditions: an initial denaturation step at 95°C for 60 seconds, followed by cycling conditions of denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds, repeated for a total of 40 cycles. Melting curves were determined under the corresponding conditions. Each sample was subjected to triplicate experiments. The reference gene GAPDH was used for normalization. The relative expression levels of the target genes were calculated using the 2-ΔΔCt method.
Western Blot
Western Blot technique was employed to assess the protein expression levels of EIF4E, VEGF, and CCND1 in the treatment and control groups. Initially, cell samples collected using RIPA lysis buffer were lysed, and the total protein concentration was determined using the BCA assay kit (Shanghai Biyuntian Biotechnology, Product No.: P0012S). Based on the detected concentration, 20 μg of total protein was loaded per well. Electrophoresis was carried out using 5% stacking gel and 10% separating gel. Subsequently, the following primary antibodies were used for immune reactions: rabbit anti-human polyclonal antibody against phospho-EIF4E (Beijing Boao Sen Biotechnology, Product No.: bs-2446R, dilution 1:1000), mouse anti-human monoclonal antibody against EIF4E (Wuhan Sanying Biotechnology, Product No.: 66655-1-Ig, dilution 1:5000), mouse anti-human monoclonal antibody against VEGFA (Wuhan Sanying Biotechnology, Product No.: 66828-1-Ig, dilution 1:1000), mouse anti-human monoclonal antibody against CCND1 (Wuhan Sanying Biotechnology, Product No.: 60186-1-Ig, dilution 1:5000), and mouse anti-human monoclonal antibody against GAPDH (Shanghai Biyuntian Biotechnology, Product No.: AF0006, dilution 1:1000). Subsequently, secondary antibodies conjugated with horseradish peroxidase (Shanghai Biyuntian Biotechnology, Product No.: A0216, dilution 1:1000) were used for immune reactions. Finally, super-sensitive ECL chemiluminescence reagent (Shanghai Biyuntian Biotechnology, Product No.: P0018S) was employed for visualization, and the ChemiDocTM Imaging System (Bio-Rad Laboratories, USA) was used for image analysis.
Statistical Analysis
GraphPad software was used for statistical analysis. Data were presented as (x ± s) and analyzed using the t-test for quantitative data. Pearson correlation analysis was performed for assessing correlations. A significance level of P < 0.05 was considered statistically significant.
Results
3D Cell Culture of SKOV3 Cells and Inhibitory Effect of 4EGI-1 on Aggregation
In this experiment, SKOV3 cells were subjected to 3D cell culture, and the impact of the EIF4E inhibitor 4EGI-1 on ovarian cancer cell aggregation was investigated. As depicted in Figure 1, compared to the control group (Figure 1A), the diameter of the SKOV3 cell spheres significantly decreased in the treatment group (Figure 1B) when exposed to 4EGI-1 under identical culture conditions. This observation indicates that inhibiting EIF4E expression effectively suppresses tumor aggregation.
Expression and Correlation Analysis of VEGFA, CCND1, and EIF4E in Ovarian Cancer Samples
To investigate the expression of VEGFA, CCND1, and EIF4E in ovarian cancer, we utilized the GEPIA online analysis tool and employed the Pearson correlation analysis method to compare expression differences between tumor and normal groups. As depicted in Figures 2A-C, the results indicate significantly elevated expression levels of VEGFA, CCND1, and EIF4E in the tumor group compared to the normal control group. Notably, the expression differences of VEGFA and CCND1 were statistically significant (p < 0.05). Furthermore, the correlation analysis revealed a positive correlation between VEGFA and CCND1 with EIF4E (Figures 2D-E), and this correlation exhibited significant statistical differences (p < 0.001). These findings suggest a potential pivotal role of VEGFA, CCND1, and EIF4E in the initiation and progression of ovarian cancer, indicating the presence of intricate interrelationships among them.
EIF4E, VEGFA, and CCND1 mRNA Expression in SKOV3 Cells
To investigate the function of EIF4E in SKOV3 cells, we conducted RT-PCR experiments comparing EIF4E inhibition group with the control group. As illustrated in Figure 3, treatment with 4EGI-1 significantly reduced EIF4E expression (0.58±0.09 vs. control, p < 0.01). Concurrently, mRNA expression of VEGFA (0.76±0.15 vs. control, p < 0.05) and CCND1 (0.81±0.11 vs. control, p < 0.05) also displayed a substantial decrease. These findings underscore the significant impact of EIF4E inhibition on the expression of VEGFA and CCND1, indicating statistically significant differences.
Protein Expression Profiles in SKOV3 Cells with EIF4E Inhibition and Control Group
Protein expression of EIF4E, VEGFA, and CCND1 was assessed using Western Blot in the 4EGI-1 treatment group and the control group. As presented in Figure 4, the expression of p-EIF4E was significantly lower in the 4EGI-1 treatment group compared to the control group (0.33±0.14 vs. control, p < 0.001). Simultaneously, the expression of VEGFA (0.53±0.18 vs. control, p < 0.01) and CCND1 (0.44±0.16 vs. control, p < 0.001) in the 4EGI-1 treatment group exhibited a marked reduction compared to the control group.
Discussion
EIF4E is a post-transcriptional modification factor that plays a pivotal role in protein synthesis. Recent studies have underscored its critical involvement in various cancers [10]. In the context of ovarian cancer research, elevated EIF4E expression has been observed in late-stage ovarian cancer tissues, with low EIF4E expression correlating to higher survival rates [9]. Suppression of EIF4E expression or function has been shown to inhibit ovarian cancer cell proliferation, invasion, and promote apoptosis. Various compounds and drugs that inhibit EIF4E have been identified, rendering them potential candidates for ovarian cancer treatment [11]. Based on the progressing understanding of EIF4E's role in ovarian cancer, inhibiting EIF4E has emerged as a novel therapeutic avenue for the disease. 4EGI-1, a cap-dependent translation small molecule inhibitor, has been suggested to disrupt the formation of the eIF4E complex [12]. In this study, our analysis of public databases revealed elevated EIF4E expression in ovarian cancer patients compared to normal controls. Furthermore, through treatment with 4EGI-1 in the SKOV3 ovarian cancer cell line, we observed a capacity for 4EGI-1 to inhibit SKOV3 cell spheroid formation. Concurrently, results from PCR and Western Blot analyses demonstrated effective EIF4E inhibition by 4EGI-1. Collectively, 4EGI-1 effectively suppresses EIF4E expression and may exert its effects on ovarian cancer therapy by modulating EIF4E.
Vascular Endothelial Growth Factor (VEGF) is a protein that stimulates angiogenesis and increases vascular permeability, playing a crucial role in tumor growth and metastasis [13]. In ovarian cancer, excessive release of VEGF by tumor cells leads to increased angiogenesis, forming a new vascular network to provide nutrients and oxygen to tumor cells. The formation of new blood vessels enables tumor growth, proliferation, and facilitates tumor cell dissemination into the bloodstream, contributing to distant metastasis [14]. As a significant member of the VEGF family, VEGFA has been extensively studied, and it has been reported that VEGFA expression is notably higher in ovarian cancer tumors [15], consistent with our public database analysis. Furthermore, elevated EIF4E levels have been associated with increased malignant tumor VEGF mRNA translation [16]. Through the use of the EIF4E inhibitor 4EGI-1 in ovarian cancer cell lines, we observed a downregulation in both mRNA and protein expression levels of VEGFA. This suggests that EIF4E inhibition might affect ovarian cancer cell angiogenesis capability through downregulation of VEGF expression.
Cyclin D1 (CCND1) is a cell cycle regulatory protein that participates in controlling cell entry into the S phase and the cell division process. In ovarian cancer, overexpression of CCND1 is associated with increased tumor proliferation activity and poor prognosis [17]. Elevated CCND1 levels promote cell cycle progression, leading to uncontrolled cell proliferation [18]. Additionally, CCND1 can activate cell cycle-related signaling pathways, promoting cancer cell growth and invasion capabilities [19]. Studies have shown that CCND1 gene expression is significantly higher in ovarian cancer tissues compared to normal ovarian tissues [20], potentially promoting proliferation and cell cycle progression through enhanced cyclin D1 translation [9]. Our public database analysis results confirm these observations. Furthermore, treatment with the EIF4E inhibitor 4EGI-1 in ovarian cancer cell lines resulted in varying degrees of downregulation in CCND1 mRNA and protein levels. This indicates that EIF4E inhibition might affect ovarian cancer cell proliferation and cell cycle progression through regulation of CCND1 expression.
In conclusion, overexpression of EIF4E appears to be closely associated with the clinical and pathological characteristics of ovarian cancer patients. In various tumors, EIF4E is significantly correlated with VEGF and cyclin D1, suggesting its role in the regulation of protein translation related to angiogenesis and growth [9, 21]. The correlation analysis results in our study further confirmed the positive correlation among EIF4E, VEGFA, and CCND1 in ovarian cancer. Simultaneous inhibition of EIF4E also led to downregulation of VEGFA and CCND1 expression, validating their interconnectedness. Thus, targeted therapy against EIF4E may prove to be an effective strategy for treating ovarian cancer. However, further research and clinical trials are necessary to assess the safety and efficacy of targeted EIF4E therapy, offering more effective treatment options for ovarian cancer patients.
Acknowledgments:
Funding: This study was supported by the Joint Project of Southwest Medical University and the Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University (Grant No. 2020XYLH-043).
Conflict of Interest: The authors declare no conflicts of interest.
#Ovarian cancer#Eukaryotic translation initiation factor 4E#Vascular endothelial growth factor A#Cyclin D1#Review Article in Journal of Clinical Case Reports Medical Images and Health Sciences .#jcrmhs
2 notes
·
View notes
Text
Activated Carbon Industry Analysis in GCC: Activated Carbon Industry Analysis in GCC Size, Growth Projections, and Competitive Landscape Analysis 2034
A new study by Fact.MR estimates that the demand for activated carbon in the Gulf Cooperation Council (GCC) will reach a market value of US$ 134.5 million in 2024. Sales are expected to grow at a CAGR of 7.6%, reaching US$ 257.5 million by 2034.
Air purification and water treatment plants in the GCC are projected to be key consumers of activated carbon, which is essential for improving odor and removing impurities from water. Additionally, the presence of a robust oil and gas industry is anticipated to drive further demand for activated carbon in the region.
𝐃𝐨𝐰𝐧𝐥𝐨𝐚𝐝 𝐚 ��𝐚𝐦𝐩𝐥𝐞 𝐂𝐨𝐩𝐲 𝐨𝐟 𝐓𝐡𝐢𝐬 𝐑𝐞𝐩𝐨𝐫𝐭: https://www.factmr.com/connectus/sample?flag=S&rep_id=9434
Country-wise Insights
The manufacturing sector is a key contributor to the United Arab Emirates' economy, with industries such as plastics & rubber, minerals, chemicals, and food & beverages expected to account for a significant portion of this sector. Moreover, the UAE's manufacturing sector is projected to see substantial growth in the coming years.
The ease of establishing and maintaining supply chain clusters is anticipated to boost the demand for activated carbon. Additionally, the increasing use of activated carbon across various industries, including air purification, water treatment, and food & beverages, is expected to create new revenue opportunities.
Read More: https://www.factmr.com/report/activated-carbon-industry-analysis-in-gcc
Category-wise Insights
The liquid form of activated carbon is expected to account for 54.3% of the regional market share by the end of 2034. Its ease of application and integration into existing systems makes it more versatile compared to solid forms. Liquid activated carbon can be sprayed, pumped, or mixed with water, offering greater flexibility in usage.
In liquid form, activated carbon particles disperse more effectively, increasing contact with substances. This improved dispersion enhances its ability to adsorb contaminants, impurities, or toxins from air, water, and other solutions, making it a more efficient option for purification processes.
𝐆𝐞𝐭 𝐂𝐮𝐬𝐭𝐨𝐦𝐢𝐳𝐚𝐭𝐢𝐨𝐧 𝐨𝐧 𝐭𝐡𝐢𝐬 𝐑𝐞𝐩𝐨𝐫𝐭 𝐟𝐨𝐫 𝐒𝐩𝐞𝐜𝐢𝐟𝐢𝐜 𝐑𝐞𝐬𝐞𝐚𝐫𝐜𝐡 𝐒𝐨𝐥𝐮𝐭𝐢𝐨𝐧𝐬: https://www.factmr.com/connectus/sample?flag=RC&rep_id=9434
Competition Landscape
Key suppliers of activated carbon are spending on new development, management of their supply chain, quality control, etc. Cabot Corporation, JACOBI CARBONS GROUP, Donau Carbon GmbH, Calgon Carbon Corporation, NTC Dubai, Fujairah Chemical, DUBI CHEM Marine International, Carbon Activated Corporation, and Veolia Water SA are prominent players in the market in GCC.
Segmentation of Activated Carbon Study in GCC
By Form :
Liquid
Solid
By Application :
Liquid Phase
Gas Phase
By Country :
Kingdom of Saudi Arabia
United Arab Emirates
Kuwait
Qatar
Oman
Bahrain
𝐂𝐨𝐧𝐭𝐚𝐜𝐭:
US Sales Office 11140 Rockville Pike Suite 400 Rockville, MD 20852 United States Tel: +1 (628) 251-1583, +353-1-4434-232 Email: [email protected]
1 note
·
View note
Text
Optimizing Market Research: Sampling Strategies by Tehrihills Consulting

Introduction -
Sampling is a fundamental aspect of market research, allowing researchers to gain insights into a larger population without having to study each individual within it. At Tehrihills Consulting, we understand the critical role of sampling in facilitating accurate and actionable Market Data Analysis. Our approach integrates advanced techniques to ensure the representativeness and reliability of the data collected in global market research.
Imagine a scenario where a company aims to understand consumer preferences across different demographics for a new product launch. Conducting surveys or studies with the entire population would be impractical and costly. Instead, through strategic sampling, we can select representative samples from each demographic group and analyze their preferences, behaviors, and opinions. This approach not only streamlines the research process but also provides valuable insights that drive informed decision-making.
Sampling is not just about selecting a subset of the population; it's about strategically choosing the right sample to represent the entire population accurately. By leveraging advanced sampling techniques and cutting-edge research methodologies, Tehrihills Consulting ensures that our clients receive accurate, reliable, and actionable insights to fuel their business growth through global data collection and analysis.
Types of sampling: sampling methods -
1. Probability Sampling Methods: Researcher employ specific criteria to randomly select members from a population. Each member within the population has an equal chance of being chosen for inclusion in the sample, ensuring fairness and representativeness. This systematic approach allows Tehrihills Consulting to gather diverse insights from various segments of the target population, driving robust market research and data analysis outcomes.
Simple Random Sampling: Simple random sampling involves selecting elements from a population entirely at random, ensuring each member has an equal chance of being chosen. This method, akin to picking a name out of a hat, offers an unbiased approach to sample selection. For instance, in an organization of 300 employees, each individual has an equal opportunity of being selected for a sample, facilitating comprehensive insights for Tehrihills Consulting's global market research initiatives.
Systematic Sampling: Systematic sampling entails selecting elements at regular intervals from a population after an initial random selection. This method offers flexibility in sample selection intervals, allowing researchers to avoid unintentional clustering. For example, a researcher may choose every 10th individual from a population of 7000 to form a systematic sample, optimizing time and resources for data collection endeavors in global data collection and analysis
Stratified Sampling: Stratified sampling involves dividing a population into distinct groups based on predefined criteria and then selecting samples randomly from each group. This method enables researchers to obtain representative samples from diverse segments of the population. For instance, Tehrihills Consulting may stratify a population based on income levels to analyse consumer behaviours across various income brackets, thereby enhancing the efficacy of Market Data Analysis.
Cluster Sampling: Cluster sampling entails selecting entire groups or clusters from a population rather than individual units. These clusters, such as geographic regions or academic cohorts, are randomly chosen to represent the entire population. By organizing clusters based on demographic parameters, Tehrihills Consulting can derive actionable insights from diverse population segments, facilitating targeted marketing strategies and informed decision-making in global market research.
2. Non-Probability Sampling Methods: The researchers handpick sample participants according to the specific objectives of their research goals. Unlike probability-based approaches, this method lacks a rigid or predetermined selection process, resulting in varying opportunities for different elements of the population to be part of the sample. This flexible methodology enables Tehrihills Consulting to tailor sampling techniques to meet the unique requirements of each research project, ensuring comprehensive and targeted insights for our clients.
Convenience Sampling: Convenience sampling involves selecting samples based on their accessibility and proximity to the researcher. While convenient, this method may introduce bias due to its non-random nature. However, it serves as a pragmatic approach in situations with time and resource constraints, allowing Tehrihills Consulting to gather preliminary insights swiftly.
Quota Sampling: Quota sampling involves setting predetermined quotas for sample attributes, such as gender or age, to ensure representation across target demographics. While quota sampling facilitates demographic diversity in samples, Tehrihills Consulting acknowledges the potential for bias in sample selection and strives to mitigate it through rigorous research market research and data analysis methodologies.
Purposive Sampling: Purposive sampling entails selecting samples based on the researcher's judgment and expertise, focusing on specific characteristics relevant to the research objectives. While purposive sampling may not yield representative samples, it offers a quick and efficient means of obtaining insights tailored to client's market research solutions needs.
Snowball Sampling: Snowball sampling involves recruiting participants through referrals from existing sample members, cascading through interconnected networks like a snowball rolling downhill. This method proves invaluable in reaching elusive populations or addressing sensitive topics, ensuring Tehrihills Consulting's research initiatives encompass diverse perspectives.

Conclusion -
In conclusion, sampling methods serve as the bedrock of market research, enabling Tehrihills Consulting to gather comprehensive data for Global data collection and analysis and Market Data Analysis. By employing a diverse array of sampling techniques, we ensure the accuracy, reliability, and relevance of insights that drive our clients' success. Partner with Tehrihills Consulting for unparalleled market research solutions tailored to your business objectives.
Unlock the power of Market Data Analysis with Tehrihills Consulting – your trusted partner for Market Research and Data Analysis. Explore global data collection and analysis, Conjoint Data Analysis, Decipher survey programming, CATI Market Research & MaxDiff Analysis. Trust Tehrihills Consulting for accurate Global Market Research Analytics, and expert Data Validation. Partner with us for comprehensive market research solutions that leverage advanced Market Data Analysis techniques to propel your business forward.
#market research#survey programming#tehrihillsconsulting#decipher#forsta#survey#researchreports#marketresearch#surveyprogramming#marketresearchsolutions#datavalidation#MaxDiffanalysis#CATImarketresearch#dataanalysis#MarketInsights#customizedresearch#customerinsights
2 notes
·
View notes
Text
Reference saved in our archive
Another great reason to mask: Covid can infect many other animals, and their mutated covid strains can reinfect us too. Keep everyone safe and healty: Mask up in public.
Abstract Background The pandemic caused by SARS-CoV-2 has not only affected humans but also raised concerns about its transmission to wild animals, potentially creating natural reservoirs. Understanding these dynamics is critical for preventing future pandemics and developing control strategies. This study aims to investigate the presence of SARS-CoV-2 in wild mammals at the Belo Horizonte Zoo in Brazil, analyzing the virus's evolution and zoonotic potential.
Methods The study was conducted at the Belo Horizonte Zoo, Minas Gerais, Brazil, covering a diverse population of mammals. Oropharyngeal, rectal, and nasal swabs were collected from 47 captive animals between November 2021 and March 2023. SARS-CoV-2 presence was determined using RT-PCR, and positive samples were sequenced for phylogenetic analysis. Consensus genomes were classified using Pangolin and NextClade tools, and a maximum likelihood phylogeny was inferred using IQ-Tree.
Results Of the 47 animals tested, nine (19.1%) were positive for SARS-CoV-2. Positive samples included rectal, oropharyngeal, and nasal swabs, with the highest positivity in rectal samples. Three genomes were successfully sequenced, revealing two variants: VOC Alpha in a maned wolf (Chrysocyon brachyurus) and a fallow deer (Dama dama), and VOC Omicron in a western lowland gorilla (Gorilla gorilla gorilla). Phylogenetic analysis indicated potential human-to-animal transmission, with animal genomes clustering close to human samples from the same region.
Conclusions This study highlights the presence of SARS-CoV-2 in various wild mammal species at the Belo Horizonte Zoo, emphasizing the virus's zoonotic potential and the complexity of interspecies transmission. The detection of different variants suggests ongoing viral evolution and adaptation in new hosts. Continuous monitoring and genomic surveillance of SARS-CoV-2 in wildlife are essential for understanding its transmission dynamics and preventing future zoonotic outbreaks. These findings underscore the need for integrated public health strategies that include wildlife monitoring to mitigate the risks posed by emerging infectious diseases.
#mask up#public health#wear a mask#pandemic#covid#wear a respirator#covid 19#still coviding#coronavirus#sars cov 2
31 notes
·
View notes
Text

(CNN) — Egypt’s Great Pyramid and other ancient monuments at Giza exist on an isolated strip of land at the edge of the Sahara Desert.
The inhospitable location has long puzzled archaeologists, some of whom had found evidence that the Nile River once flowed near these pyramids in some capacity, facilitating the landmarks’ construction starting 4,700 years ago.
Using satellite imaging and analysis of cores of sediment, a new study published Thursday in the journal Communications Earth & Environment has mapped a 64-kilometer (40-mile) long, dried-up branch of the Nile, long buried beneath farmland and desert.
“Even though many efforts to reconstruct the early Nile waterways have been conducted, they have largely been confined to soil sample collections from small sites, which has led to the mapping of only fragmented sections of the ancient Nile channel systems,” said lead study author Eman Ghoneim, a professor and director of the Space and Drone Remote Sensing Lab at the University of North Carolina Wilmington’s Department of Earth and Ocean Sciences.
“This is the first study to provide the first map of the long-lost ancient branch of the Nile River.”

Ghoneim and her colleagues refer to this extinct branch of the Nile river as Ahramat, which is Arabic for pyramids.
The ancient waterway would have been about 0.5 kilometers wide (about one-third of a mile) with a depth of at least 25 meters (82 feet) — similar to the contemporary Nile, Ghoneim said.
“The large size and extended length of the Ahramat Branch and its proximity to the 31 pyramids in the study area strongly suggests a functional waterway of great importance,” Ghoneim said.
She said the river would have played a key role in ancient Egyptians’ transportation of the enormous amount of building materials and laborers needed for the pyramids’ construction.
“Also, our research shows that many of the pyramids in the study area have (a) causeway, a ceremonial raised walkway, that runs perpendicular to the course of the Ahramat Branch and terminates directly on its riverbank.”
Hidden traces of a lost waterway

Traces of the river aren’t visible in aerial photos or in imagery from optical satellites, Ghoneim said.
In fact, she only spotted something unexpected while studying radar satellite data of the wider area for ancient rivers and lakes that might reveal a new source of groundwater.
“I am a geomorphologist, a paleohydrologist looking into landforms. I have this kind of trained eye,” she said.
“While working with this data, I noticed this really obvious branch or a kind of riverbank, and it didn’t make any sense because it is really far from the Nile,” she added.
Born and raised in Egypt, Ghoneim was familiar with the cluster of pyramids in this area and had always wondered why they were built there.
She applied to the National Science Foundation to investigate further.
Geophysical data taken at ground level with the use of ground-penetrating radar and electromagnetic tomography confirmed it was an ancient arm of the Nile.
Two long cores of earth the team extracted using drilling equipment revealed sandy sediment consistent with a river channel at a depth of about 25 meters (82 feet).
It’s possible that “countless” temples might still be buried beneath the agricultural fields and desert sands along the riverbank of the Ahramat Branch, according to the study.

Why this branch of the river dried up or disappeared is still unclear. Most likely, a period of drought and desertification swept sand into the region, silting up the river, Ghoneim said.
"The study demonstrated that when the pyramids were built, the geography and riverscapes of the Nile differed significantly from those of today," said Nick Marriner, a geographer at the French National Centre for Scientific Research in Paris.
He was not involved in the study but has conducted research on the fluvial history of Giza.
“The study completes an important part of the past landscape puzzle,” Marriner said.
“By putting together these pieces, we can gain a clearer picture of what the Nile floodplain looked like at the time of the pyramid builders and how the ancient Egyptians harnessed their environments to transport building materials for their monumental construction endeavors.”

#Egypt#Great Pyramid#Giza#Nile River#archaeology#Ancient Egypt#Communications Earth & Environment#Space and Drone Remote Sensing Lab#map#pyramids#Ahramat#ground-penetrating radar#electromagnetic tomography#Unas#Valley Temple#Great Sphinx of Giza#pyramid mystery#waterways
6 notes
·
View notes
Text

Hubble Space telescope unveils the first images of ongoing star cluster mergers near the center of dwarf galaxies
A new study reports the first direct observation of merging star clusters in the nuclear region of dwarf galaxies. This detection confirms the feasibility of this formation route for nuclei in dwarf galaxies, which has long been debated. The study was published in Nature science journal, and led by Postdoctoral Researcher Mélina Poulain from the University of Oulu, Finland.
Dwarf galaxies are the most abundant type of galaxies that populate the Universe. Composed of 100 times fewer stars than the Milky Way, or even less, they are the building blocks of more massive galaxies. Thus, understanding their formation is key to comprehend galaxy evolution.
A notable fraction of dwarf galaxies host a compact star cluster at their centers, typically made of hundreds of thousands to hundreds of millions of stars. Known as nuclear star clusters, these are the densest type of stellar systems in the Universe. The formation of such extreme objects has been under debate for several decades. In dwarf galaxies, they are believed to form from the merger of smaller star clusters, called globular clusters, after they migrate to the galaxy center. However, no such merger of globular clusters has been directly observed to confirm the theory, until now.
Witnessing rare features
While studying observations of a large sample of nearly 80 dwarf galaxies from the Hubble Space telescope, which were led by Prof. Francine Marleau at the University of Innsbruck, Austria, a group of ten researchers from the international MATLAS collaboration noticed a handful of galaxies with an unusual looking nuclear star cluster. Some showed a couple of star clusters close together, while others had a feature similar to a faint stream of light attached to the nuclear star cluster.
“We were surprised by the streams of light that were visible near the center of the galaxies, as nothing similar has been observed in the past”, explains Mélina Poulain.
A thorough analysis of the features has shown that they have similar properties to globular clusters already detected in dwarf galaxies. This suggests that the observations witness the growth of the nuclear star cluster by the dramatic cannibalization of globular clusters at the cores of those galaxies.
Observations reproduced in simulations
To confirm the origin of the faint streams of light, ultra-high resolution complementary simulations were implemented to model the merging process. This portion of the work, led by Dr. Rory Smith at the Universidad Técnica Federico Santa María in Santiago, Chile, set up various mergers between star clusters with differing masses, dynamics, and numbers of clusters involved. Results confirm that the observed light streams are created with two star clusters with significant mass differences merge. The larger the mass ratio, the longer the stream. The process typically lasts a short amount of time, less than 100 million years, and the features produced are visible for even less time, which explains the difficulty of catching such a phenomenon.
2 notes
·
View notes
Text
Researchers Unveil New Disease-Linked Microbial Species through Pan-Body Microbiome Analysis
https://thelifesciencesmagazine.com/wp-content/uploads/2024/10/TLM-1-10-2024-1-Researchers-Unveil-New-Disease-Linked-Microbial-Species-through-Pan-Body-Microbiome-Analysis-Source-bacdive.dsmz_.de_.jpg
[Source – bacdive.dsmz.de]
A recent study conducted by researchers in Germany and published in Nature Communications has revealed new insights into the human microbiome’s role in disease diagnosis and management. Through a comprehensive pan-body microbiome analysis, the team examined multiple body sites across various diseases, uncovering key microbial species linked to specific health conditions. This approach highlights the importance of understanding the microbiome’s complexity across the human body, offering new avenues for diagnostic and therapeutic developments.
The Pan-Body Microbiome Approach
The human microbiome, a diverse community of microorganisms living in and on the body, varies across different regions and plays a crucial role in health and disease. This variability has long been studied in relation to specific organs, but the pan-body approach adopted in this study takes a broader view. By examining the microbiota of multiple organ systems, the researchers aimed to uncover a more holistic understanding of disease mechanisms.
From 2021 to 2023, researchers collected 1,931 high-quality samples from 515 patients suffering from chronic inflammatory conditions affecting the lungs, eyes, oral cavity, heart, skin, and intestines. These samples, including dental plaque, saliva, skin, stool, and eye specimens, underwent advanced sequencing techniques, which helped identify microbial imbalances and potential links to diseases. Their findings showed that alterations in the microbiome are often associated with the onset and progression of diseases, with certain species being more prevalent in specific conditions.
Key Findings and Potential Therapeutic Applications
The study uncovered 583 previously unexplored species genome bins (SGBs), 189 of which were strongly associated with diseases. Oral microbiome samples, such as saliva and dental plaque, exhibited a higher degree of novelty, accounting for 72% of newly identified species. For instance, Corynebacterium pseudogenitalium and Staphylococcus epidermidis were found in higher abundance on the skin of patients with coronary heart disease, while specific microbial species were linked to conditions like obesity, digestive diseases, and Parkinson’s disease.
In addition to species identification, the researchers discovered a significant association between antimicrobial resistance (AMR) genes and certain diseases. Notably, resistance genes such as New Delhi metallo beta-lactamases (NDM) and oxacillin-hydrolyzing carbapenemases (OXA) were prevalent across samples, highlighting the spread of drug resistance in various microbial communities. The study also found that microbial compositions varied depending on dietary habits, with certain species more abundant in omnivorous participants compared to vegetarians.
Importantly, the researchers developed a novel method for prioritizing biosynthetic gene clusters (BGCs) with high therapeutic potential. These BGCs, which produce compounds similar to known antibiotics, could serve as a foundation for developing new drugs. This discovery is particularly significant for addressing the growing concern of antibiotic resistance.
Implications for Future Research and Medicine
The study’s findings underscore the importance of a pan-body, pan-disease microbiome approach in diagnosing and managing diseases. By investigating multiple body sites simultaneously, researchers were able to identify novel microbial species and their potential therapeutic applications. The results call for further research into the functionality of these newly discovered BGCs, particularly in terms of their biotechnological suitability and potential for drug development.
This groundbreaking research paves the way for future medical discoveries, particularly in the development of new antibiotics, and highlights the need for a more integrated approach to studying the human microbiome in relation to disease.
1 note
·
View note