#Single-cell RNA Data
Explore tagged Tumblr posts
Text
0 notes
Text
the whole question of whether viruses are or are not really "alive" is kind of interesting to me because viruses are kind of like software if you think about it?
Like a virus on its own is a fairly inert thing - it's a packet of RNA code inside a protein casing and doesn't really do anything on its own except float around until they contact a matching cell membrane whereupon the casing gets hooked up and the RNA injected. In software terms, RNA is kind of like a segment of free-floating biological machine code, while the casing is the encapsulating data structure that allows it to interface with things, like the MZ EXE format of the DOS/Windows world or the ELF format of Unix and its relatives. What effectively makes biological viruses work at all is the fact that cells tend to absorb the contents of anything that can bind to their surface and have minimal if any protections in place against processing foreign RNA code. In computer security terms, cells are extremely vulnerable to remote code execution, and the antiviral protections that the body has is primarily a question of either identifying and eliminating viral particles before they get linked up to a cell, or murdering any cell that looks infected.
Okay that's interesting but what does that have to do with whether or not viruses are alive you may ask, so let me pull this analogy together a little by asking the following: Where does software exist?
This might seem like a silly question at first, but it's actually not as simple as it might seem. Consider this post you're reading - where is it? Well, on the servers of tumblr dot com you may say, but you're not looking at the servers right now, are you. Okay well a local copy on the device you're reading this on too then - and sure, there is definitely such a copy, but you're not looking at that either, not directly at least: that data only exists in memory as electrical signals and charges on a few microchips are not something we can see either. No, what you're looking at is an image most likely rendered on a screen through a complex interplay of code and data, of both hardware and software operating together, and only from that full stack of interoperating elements does the post you're reading emerge in a form that you can read.
Or, to go back to the main topic: viruses are code - code which comes alive when introduced to a living cell, but lies inert within the virus itself. They live in the sense that they have functional biological machine code, if you will, but lack the active process with which to execute said code themselves. Like software needs some kind of computer to run, a virus needs a living cell to run - they are alive but only when inside a cell, on their own in isolation they are just inert clumps of protein that depend on being picked up and executed.
A virus is kind of the ultimate expression of life as an emergent property: no one single part of the virus itself is really alive, and yet when introduced to a susceptible cell, its code will interact with the processes of that cell and real, living behaviour emerges. The life of viruses is not contained anywhere within the virus itself, it is something that only exists in interaction with other things outside of itself.
7 notes
·
View notes
Text

The first ancestor | O primeiro ancestral
🇬🇧
The origin of life on Earth is a fascinating and complex topic that continues to be the subject of scientific investigation and debate. The exact details of how and when life first emerged are not yet fully understood, but there are several theories and hypotheses.
One prominent theory suggests that life on Earth originated in hydrothermal vents at the ocean floor. These environments provided a rich mix of chemicals and minerals that could have supported the formation of simple organic molecules. Over time, these molecules might have developed into more complex structures, leading to the first self-replicating entities. Another hypothesis proposes that life's “building blocks” may have been delivered to Earth through comets or asteroids, carrying organic molecules from space. This concept, known as panspermia, suggests that the raw materials for life could have arrived on Earth from extraterrestrial sources.
The transition from simple organic molecules to the first living organisms is a critical step in the emergence of life. The earliest life forms were likely simple, single-celled organisms. These early life forms, often referred to as prokaryotes, lacked a distinct nucleus and membrane-bound organelles. They were likely similar to modern-day bacteria and archaea. One crucial aspect of early life was the development of a self-replicating system, allowing for the transmission of genetic information from one generation to the next. The formation of simple genetic material, possibly in the form of RNA, played a pivotal role in this process.
The study of extremophiles beings, organisms that thrive in extreme environments such as hot springs and deep-sea hydrothermal vents, has also provided valuable data into the conditions that early life might have faced. While the details remain a subject of ongoing research, understanding the origins of life on Earth is crucial for unraveling the broader story of our planet's history and the potential for life elsewhere in the universe. Scientists continue to explore these questions through various disciplines, including paleontology, molecular biology, and astrobiology.
🇧🇷
A origem da vida na Terra é um tópico fascinante e complexo que continua a ser objeto de investigação científica e debate. Os detalhes exatos de como e quando a vida surgiu ainda não são completamente compreendidos, mas existem várias teorias e hipóteses.
Uma teoria proeminente sugere que a vida na Terra teve origem em fontes hidrotermais no fundo do oceano. Esses ambientes forneciam uma mistura rica de produtos químicos e minerais que poderiam ter apoiado a formação de moléculas orgânicas simples. Com o tempo, essas moléculas poderiam ter se desenvolvido em estruturas mais complexas, levando aos primeiros seres auto-replicantes. Outra hipótese propõe que os “blocos de construção da vida” podem ter sido entregues à Terra por meio de cometas ou asteroides, carregando moléculas orgânicas do espaço. Esse conceito, conhecido como panspermia, sugere que os materiais para a vida podem ter chegado à Terra a partir de fontes extraterrestres.
A transição de moléculas orgânicas simples para os primeiros organismos vivos é um passo crítico no surgimento da vida. Os primeiros seres vivos provavelmente eram organismos unicelulares simples. Essas formas de vida iniciais, frequentemente chamadas de procariontes, não possuíam um núcleo distinto e organelas membranosas. Eles eram provavelmente semelhantes às bactérias e arqueias modernas. Um aspecto crucial da vida inicial foi o desenvolvimento de um sistema autorreplicante, permitindo a transmissão de informações genéticas de uma geração para outra. A formação de material genético simples, possivelmente na forma de RNA, desempenhou um papel fundamental nesse processo.
O estudo de seres extremófilos, organismos que prosperam em ambientes extremos como nascentes termais e fontes hidrotermais profundas, também forneceu insights valiosos sobre as condições que a vida inicial pode ter enfrentado. Embora os detalhes permaneçam um tema de pesquisa em andamento, compreender as origens da vida na Terra é crucial para desvendar a história mais ampla de nosso planeta e o potencial de vida em outros lugares do universo. Cientistas continuam a explorar essas questões por meio de várias disciplinas, incluindo paleontologia, biologia molecular e astrobiologia.
#space#universe#paleobotany#paleontology#science#digital painting#dinosaur#life#geology#biology#biodiversity#molecular biology#dna#rna
6 notes
·
View notes
Text
Spatiotemporal Omics Market: Growth Trends, Technologies, and Future Forecast
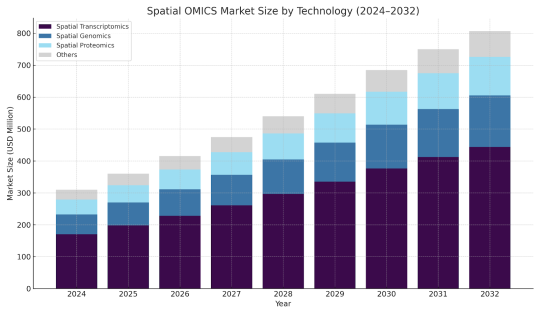
What is spatiotemporal omics?
Spatiotemporal omics is an advanced approach in molecular biology that integrates spatial and temporal dimensions into multi-omics analyses (e.g., genomics, transcriptomics, proteomics, metabolomics). This technique enables the mapping of biomolecular changes within the precise anatomical context of tissues over time, offering unprecedented resolution into how cellular behavior evolves in health and disease. It has transformative potential in areas such as oncology, neuroscience, immunology, and developmental biology, driving innovation in precision medicine and systems biology.
The Spatio OMICS Market is expected to grow at a significant rate due to advancements in sequencing and imaging technologies, and expansion of research and development funding.
Which technologies are driving the spatiotemporal omics market?
Spatial Transcriptomics – Maps gene expression in tissue context
Spatial Proteomics – Visualizes protein distribution
Mass Spectrometry Imaging (MSI) – Detects molecules with spatial precision
Single-Cell RNA Sequencing (scRNA-seq) – Captures temporal changes at cell level
Multiplexed Imaging (e.g., CODEX, MIBI) – Analyzes many biomarkers in tissues
What are the current limitations or challenges in spatiotemporal omics adoption?
High Technology Costs: The advanced instruments and reagents required for spatiotemporal omics are costly, making adoption challenging for many academic and smaller research institutions. This financial barrier limits access despite rising interest in spatial biology.
Complexity of Data Analysis: Spatiotemporal omics generate vast, high-dimensional datasets combining molecular and imaging data. Processing this information demands specialized software, computational infrastructure, and bioinformatics expertise. Without these, deriving actionable insights can be slow and resource-intensive.
Limited Skilled Workforce and Infrastructure: The field requires interdisciplinary skills in molecular biology, spatial imaging, and data science. However, a shortage of trained professionals and inadequate infrastructure in many regions slows down adoption and implementation across research and clinical environments.
To get detailed information on Spatiotemporal OMICS Industry, Click here!
Which regions are investing heavily in spatiotemporal omics research and development?
North America
Europe
Asia-Pacific
Latin America
Who are the leading players in the spatiotemporal omics industry?
10x Genomics
NanoString Technologies
Akoya Biosciences
Bruker Corporation
Vizgen
RareCyte
For a comprehensive analysis, refer to the full report by BIS Research: Spatiotemporal OMICS Market.
End Use Insights
Innovation Strategy: It identifies opportunities for market entry and technology adoption, helping organizations stay ahead of the competition while meeting evolving customer demands.
Growth Strategy: The report outlines targeted growth strategies to optimize market share, enhance brand presence, and drive revenue expansion.
Competitive Strategy: It evaluates key competitors and offers practical guidance for maintaining a competitive edge in a rapidly evolving market.
Conclusion
The market for spatiotemporal omics is expected to increase significantly due to growing applications in clinical and research settings, growing need for precision medicine, and technical advancements. To keep a competitive edge, major competitors in the market are always improving their product offerings, investing in R&D, and inventing. Despite obstacles like exorbitant expenses and intricate data, the amalgamation of artificial intelligence and multi-modal platforms offers significant prospects. Organizations that use these insights can take advantage of development opportunities, overcome obstacles, and set themselves up for long-term success in the ever-changing spatiotemporal omics landscape.
#Spatiotemporal Omics Market#Spatiotemporal Omics Industry#Spatiotemporal Omics Report#health#healthcare
0 notes
Text
Tumor Transcriptomics Market Size, Share, Trends, Demand, Growth and Competitive Analysis
Executive Summary Tumor Transcriptomics Market:
This international Tumor Transcriptomics Market business report includes strategic profiling of key players in the market, systematic analysis of their core competencies, and draws a competitive landscape for the market. It is the most appropriate, rational and admirable market research report provided with a devotion and comprehension of business needs. The report also estimates CAGR (compound annual growth rate) values along with its fluctuations for the definite forecast period. To understand the competitive landscape in the market, an analysis of Porter’s five forces model for the market has also been included in this market report. It all together leads to the company’s growth, by subsidizing the risk and improving the performance.
Competitive landscape in this report covers strategic profiling of key players in the market, comprehensively analyzing their core competencies, and strategies. According to this Tumor Transcriptomics Market report, the global market is anticipated to witness a moderately higher growth rate during the forecast period. This Tumor Transcriptomics Market report is structured with the clear understanding of business goals of industry and needs to bridge the gap by delivering the most appropriate and proper solutions. Businesses can confidently rely on the information mentioned in this Tumor Transcriptomics Market report as it is derived only from the important and genuine resources.
Discover the latest trends, growth opportunities, and strategic insights in our comprehensive Tumor Transcriptomics Market report. Download Full Report: https://www.databridgemarketresearch.com/reports/global-tumor-transcriptomics-market
Tumor Transcriptomics Market Overview
**Segments**
- **By Product Type**: The tumor transcriptomics market can be segmented into instruments, consumables, and services. Instruments include PCR machines, microarray equipment, and sequencing platforms. Consumables consist of reagents, RNA extraction kits, and assay kits. Services cover gene expression profiling, data analysis, and consulting services.
- **By Cancer Type**: This market segment is categorized into breast cancer, lung cancer, colorectal cancer, prostate cancer, and others. Each cancer type may require specific transcriptomic analysis for targeted therapies and personalized medicine.
- **By End-User**: The tumor transcriptomics market can be further divided into hospitals, cancer research centers, diagnostic laboratories, and pharmaceutical companies. Different end-users have varying needs for transcriptomic tools and services.
- **By Region**: Geographically, the market is segmented into North America, Europe, Asia Pacific, Latin America, and Middle East & Africa. Each region has its own set of regulations, healthcare infrastructure, and adoption rates for tumor transcriptomics technology.
**Market Players**
- **Illumina, Inc.**: One of the key players in the tumor transcriptomics market, Illumina offers sequencing platforms and related services for cancer research and diagnostics.
- **Thermo Fisher Scientific Inc.**: This company provides a wide range of consumables and instruments for tumor transcriptomics analysis, catering to the needs of researchers and healthcare professionals.
- **Agilent Technologies**: Known for its microarray platforms and assay kits, Agilent Technologies is a major player in the global tumor transcriptomics market, offering solutions for gene expression profiling.
- **QIAGEN N.V.**: QIAGEN specializes in RNA extraction kits and bioinformatics tools essential for tumor transcriptomics, enabling researchers to analyze gene expression patterns in cancer.
- **Fluidigm Corporation**: With its innovative microfluidic technology, Fluidigm Corporation offers high-throughput solutions for single-cell analysis and gene expression studies in tumors.
The global tumor transcriptomics market is witnessing significant growth due to the rising prevalence of cancer worldwide and the increasing demand for precision medicine. Advancements in transcriptomic technologies, such as next-generation sequencing and microarray analysis, have enabled researchers to study gene expression patterns in tumors with high accuracy and throughput. Key market players are investing in product development, strategic collaborations, and expansion initiatives to capitalize on the growing opportunities in this market. As personalized medicine gains momentum, the use of tumor transcriptomics for patient stratification and treatment selection is expected to drive further market growth.
Market players such as Illumina, Thermo Fisher Scientific Inc., Agilent Technologies, QIAGEN N.V., and Fluidigm Corporation are at the forefront of developing cutting-edge solutions for tumor transcriptomics. These companies offer a wide range of instruments, consumables, and services that cater to the diverse needs of hospitals, cancer research centers, diagnostic laboratories, and pharmaceutical companies. By focusing on product development and strategic collaborations, these key players are driving innovation in the market and expanding their global footprint.
In addition to technological advancements, the market is also influenced by regulatory landscapes and healthcare infrastructure in different regions. North America, Europe, Asia Pacific, Latin America, and the Middle East & Africa each have unique market dynamics that shape the adoption and growth of tumor transcriptomics technology. Market players must navigate these regional differences to effectively penetrate local markets and capitalize on the growing demand for precision medicine solutions.
Advancements in transcriptomic technologies, such as next-generation sequencing, microarray analysis, and RNA extraction kits, have revolutionized the way researchers study gene expression patterns in tumors. This enhanced accuracy and throughput have paved the way for more precise cancer treatments, driving the demand for transcriptomic analysis tools and services across different cancer types. The shift towards personalized medicine, which relies heavily on tumor transcriptomics to identify specific gene expression patterns for tailored treatment decisions, is a key trend shaping the market dynamics.
Furthermore, while technological innovation remains a key driver of market growth, regional dynamics also play a crucial role in shaping the adoption and expansion of tumor transcriptomics technology. Different regions such as North America, Europe, Asia Pacific, Latin America, and the Middle East & Africa have unique regulatory landscapes and healthcare infrastructures that impact market dynamics. Market players must navigate these regional differences effectively to tap into local markets and capitalize on the increasing demand for precision medicine solutions.
Looking ahead, the global tumor transcriptomics market is expected to maintain its upward trajectory as the emphasis on personalized medicine grows and the need for targeted therapies for different cancer types intensifies. Researchers and healthcare professionals are increasingly relying on transcriptomic analysis to gain a better understanding of cancer biology and develop innovative treatment strategies. Key market players will continue to drive innovation through strategic initiatives such as product launches, collaborations, and mergers, reinforcing their position in this competitive and rapidly evolving market landscape.
The Tumor Transcriptomics Market is highly fragmented, featuring intense competition among both global and regional players striving for market share. To explore how global trends are shaping the future of the top 10 companies in the keyword market.
Learn More Now: https://www.databridgemarketresearch.com/reports/global-tumor-transcriptomics-market/companies
DBMR Nucleus: Powering Insights, Strategy & Growth
DBMR Nucleus is a dynamic, AI-powered business intelligence platform designed to revolutionize the way organizations access and interpret market data. Developed by Data Bridge Market Research, Nucleus integrates cutting-edge analytics with intuitive dashboards to deliver real-time insights across industries. From tracking market trends and competitive landscapes to uncovering growth opportunities, the platform enables strategic decision-making backed by data-driven evidence. Whether you're a startup or an enterprise, DBMR Nucleus equips you with the tools to stay ahead of the curve and fuel long-term success.
Regional Analysis/Insights
The Tumor Transcriptomics Market is analyzed and market size insights and trends are provided by country, component, products, end use and application as referenced above.
The countries covered in the Tumor Transcriptomics Market reportare U.S., Canada and Mexico in North America, Germany, France, U.K., Netherlands, Switzerland, Belgium, Russia, Italy, Spain, Turkey, Rest of Europe in Europe, China, Japan, India, South Korea, Singapore, Malaysia, Australia, Thailand, Indonesia, Philippines, Rest of Asia-Pacific (APAC) in the Asia-Pacific (APAC), Saudi Arabia, U.A.E, South Africa, Egypt, Israel, Rest of Middle East and Africa (MEA) as a part of Middle East and Africa (MEA), Brazil, Argentina and Rest of South America as part of South America.
North America dominatesthe Tumor Transcriptomics Market because of the region's high prevalence Tumor Transcriptomics Market
Asia-Pacific is expectedto witness significant growth. Due to the focus of various established market players to expand their presence and the rising number of surgeries in this particular region.
Browse More Reports:
Global Respiratory Syncytial Virus Treatment Market Global Resorcinol Market Global Refrigerated Vending Machine Market Global Recliner Sofas Market Global Railway Operation Management Market Global Radio Frequency (RF) Components Market Global Rabies Diagnostics Market Global PVC Additives Market Global Puerperal Sepsis Treatment Market Global Pregnancy Pillow Market Global Power Inductor Market Global Powdered Seaweed Market Global Potassium carbonate Market Global Portable Fabric Canopies Market Global Polyurethane Foam Market Global Polysulfide Market Global Polyoxymethylene (POM) Market Global Polyolefin Market Global Polyethylene Terephthalate (PET) Stretch Blow Molding Machines Market Global Polyethylene (PE) Foam Film Market
About Data Bridge Market Research:
An absolute way to forecast what the future holds is to comprehend the trend today!
Data Bridge Market Research set forth itself as an unconventional and neoteric market research and consulting firm with an unparalleled level of resilience and integrated approaches. We are determined to unearth the best market opportunities and foster efficient information for your business to thrive in the market. Data Bridge endeavors to provide appropriate solutions to the complex business challenges and initiates an effortless decision-making process. Data Bridge is an aftermath of sheer wisdom and experience which was formulated and framed in the year 2015 in Pune.
Contact Us: Data Bridge Market Research US: +1 614 591 3140 UK: +44 845 154 9652 APAC : +653 1251 975 Email:- [email protected]
Tag
Tumor Transcriptomics Market Size, Tumor Transcriptomics Market Share, Tumor Transcriptomics Market Trend, Tumor Transcriptomics Market Analysis, Tumor Transcriptomics Market Report, Tumor Transcriptomics Market Growth, Latest Developments in Tumor Transcriptomics Market, Tumor Transcriptomics Market Industry Analysis, Tumor Transcriptomics Market Key Player, Tumor Transcriptomics Market Demand Analysis
0 notes
Text
IEEE Transactions on Evolutionary Computation, Volume 29, Issue 3
1) Guest Editorial Machine-Learning-Assisted Evolutionary Computation
Author(s): Rong Qu, Nelishia Pillay, Emma Hart, Manuel López-Ibáñez
Pages: 571 - 573
2) A Deep Reinforcement Learning-Assisted Multimodal Multiobjective Bilevel Optimization Method for Multirobot Task Allocation
Author(s): Yuanyuan Yu, Qirong Tang, Qingchao Jiang, Qinqin Fan
Pages: 574 - 588
3) An Iterated Greedy Algorithm With Reinforcement Learning for Distributed Hybrid Flowshop Problems With Job Merging
Author(s): Xin-Rui Tao, Quan-Ke Pan, Liang Gao
Pages: 589 - 600
4) Surrogate-Assisted Multiobjective Gene Selection for Cell Classification From Large-Scale Single-Cell RNA Sequencing Data
Author(s): Jianqing Lin, Cheng He, Hanjing Jiang, Yabing Huang, Yaochu Jin
Pages: 601 - 615
5) Dealing With Structure Constraints in Evolutionary Pareto Set Learning
Author(s): Xi Lin, Xiaoyuan Zhang, Zhiyuan Yang, Qingfu Zhang
Pages: 616 - 630
6) A Two-Population Algorithm for Large-Scale Multiobjective Optimization Based on Fitness-Aware Operator and Adaptive Environmental Selection
Author(s): Bingdong Li, Yan Zhang, Peng Yang, Xin Yao, Aimin Zhou
Pages: 631 - 645
7) Protein Structure Prediction Using a New Optimization-Based Evolutionary and Explainable Artificial Intelligence Approach
Author(s): Jun Hong, Zhi-Hui Zhan, Langchong He, Zongben Xu, Jun Zhang
Pages: 646 - 660
8) Multiobjective Mixed-Integer Quadratic Models: A Study on Mathematical Programming and Evolutionary Computation
Author(s): Ofer M. Shir, Michael Emmerich
Pages: 661 - 675
9) A Survey on Evolutionary Computation-Based Drug Discovery
Author(s): Qiyuan Yu, Qiuzhen Lin, Junkai Ji, Wei Zhou, Shan He, Zexuan Zhu, Kay Chen Tan
Pages: 676 - 696
10) Linear Subspace Surrogate Modeling for Large-Scale Expensive Single/Multiobjective Optimization
Author(s): Langchun Si, Xingyi Zhang, Ye Tian, Shangshang Yang, Limiao Zhang, Yaochu Jin
Pages: 697 - 710
11) A Classifier-Ensemble-Based Surrogate-Assisted Evolutionary Algorithm for Distributed Data-Driven Optimization
Author(s): Xiao-Qi Guo, Feng-Feng Wei, Jun Zhang, Wei-Neng Chen
Pages: 711 - 725
12) Improving the Efficiency of the Distance-Based Hypervolume Estimation Using ND-Tree
Author(s): Andrzej Jaszkiewicz, Piotr Zielniewicz
Pages: 726 - 733
13) A Cooperative Ant Colony System for Multiobjective Multirobot Task Allocation With Precedence Constraints
Author(s): Tong Qian, Xiao-Fang Liu, Yongchun Fang
Pages: 734 - 748
14) Evolutionary Trainer-Based Deep Q-Network for Dynamic Flexible Job-Shop Scheduling
Author(s): Yun Liu, Fangfang Zhang, Yanan Sun, Mengjie Zhang
Pages: 749 - 763
15) MOEA/D With Spatial–Temporal Topological Tensor Prediction for Evolutionary Dynamic Multiobjective Optimization
Author(s): Xianpeng Wang, Yumeng Zhao, Lixin Tang, Xin Yao
Pages: 764 - 778
16) A Surrogate-Assisted Evolutionary Framework for Expensive Multitask Optimization Problems
Author(s): Shenglian Tan, Yong Wang, Guangyong Sun, Tong Pang, Ke Tang
Pages: 779 - 793
17) Improved Evolutionary Multitasking Optimization Algorithm With Similarity Evaluation of Search Behavior
Author(s): Xiaolong Wu, Wei Wang, Tengfei Zhang, Honggui Han, Junfei Qiao
Pages: 794 - 808
18) Competitive Multitasking for Computational Resource Allocation in Evolutionary-Constrained Multiobjective Optimization
Author(s): Xiaoliang Chu, Fei Ming, Wenyin Gong
Pages: 809 - 821
19) Fractional Order Differential Evolution
Author(s): Kaiyu Wang, Shangce Gao, MengChu Zhou, Zhi-Hui Zhan, Jiujun Cheng
Pages: 822 - 835
20) An Interval Multiobjective Evolutionary Generation Algorithm for Product Design Change Plans in Uncertain Environments
Author(s): Rui-Zhao Zheng, Yong Zhang, Xiao-Yan Sun, Dun-Wei Gong, Xiao-Zhi Gao
Pages: 836 - 850
0 notes
Text
Why Everyone's Talking About the Next-Generation Sequencing Services Market
Next-Generation Sequencing Services Market Growth & Trends
The Next-Generation Sequencing Services Market is forecasted to reach USD 24.48 billion by 2030, expanding at a compound annual growth rate (CAGR) of 22.68% from 2024 to 2030, according to a report by Grand View Research, Inc. Advances in NGS technology have significantly improved gene sequencing efficiency, reproducibility, and cost-effectiveness, driving its adoption in clinical laboratories and hospitals. Recent innovations focus on simplifying sequencing technology and enhancing its sophistication, which has fueled its widespread use among various end-users.
Over the last five years, NGS has transitioned from research applications to clinical use, with 14 countries launching large-scale genome sequencing programs. By 2025, nearly 60 million individuals are expected to have their genomes analyzed. Traditional disease testing service providers have expanded their offerings to include sequencing-based genetic tests. For instance, ARUP Laboratories introduced new coronavirus tests in March 2022, while IDbyDNA, Inc. collaborated to develop an NGS-based test for respiratory illnesses, assisting physicians in diagnosing pneumonia and other respiratory conditions.
The increasing demand for high-throughput sequencing within the pharmaceutical and biotechnology sectors has necessitated improvements in sequencing speed, platform efficiency, and workflow simplification. Companies are now focusing on enhancing sample amplification and purification technologies. In March 2022, Illumina, Inc. unveiled TruSight Oncology (TSO) Comprehensive (EU), a diagnostic tool designed to analyze multiple tumor genes and biomarkers, enabling personalized genomic decisions for cancer patients. This in vitro diagnostic (IVD) kit debuted in Europe, marking the beginning of its global rollout.
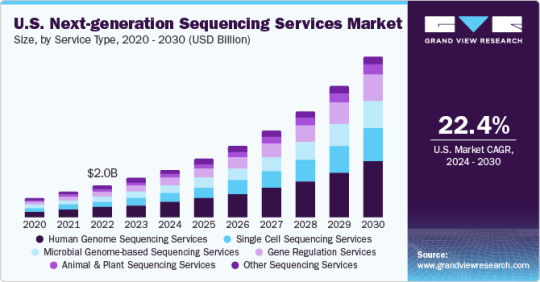
Curious about the Next-Generation Sequencing Services Market? Download your FREE sample copy now and get a sneak peek into the latest insights and trends.
Next-Generation Sequencing Services Market Report Highlights
By service type, human genome sequencing held the largest revenue share in 2023 owing to greater penetration of whole genome and whole exome sequencing
Gene regulation services, particularly small RNA sequencing and ChIP-seq, are anticipated to grow at a lucrative pace during the forecast period. Rising investment in RNA sequencing is fueling the growth of gene regulation services
The larger revenue share of sequencing services as compared to other workflow steps can be attributed to an increase in the installation of sequencing platforms. High maintenance and recovery costs of this step have also resulted in greater revenue generation
By end-use, the universities and other research entities segment held the largest revenue share in 2023 owing to the growing application of NGS technology for cancer prognosis as well as diagnosis and major projects taken up by academicians and universities
North America dominated the market in 2023 owing to higher penetration of the technology in this region and the availability of funds to employ the technique and derive research findings for disease targeting
Next-Generation Sequencing Services Market Segmentation
Grand View Research has segmented the global next-generation sequencing services market report based on service type, workflow, end-use, and region
Next-generation Sequencing (NGS) Service Type Outlook (Revenue, USD Million, 2018 - 2030)
Human Genome Sequencing Services
Single Cell Sequencing Services
Microbial Genome-based Sequencing Services
Gene Regulation Services
Small RNA Sequencing
ChIP Sequencing
Other Gene Regulation-based Services
Animal & Plant Sequencing Services
Other Sequencing Services
Next-generation Sequencing (NGS) Services Workflow Outlook (Revenue, USD Million, 2018 - 2030)
Pre-sequencing
Sequencing
Data Analysis
Next-generation Sequencing (NGS) Services End-use Outlook (Revenue, USD Million, 2018 - 2030)
Universities & Other Research Entities
Hospitals & Clinics
Pharma & Biotech Entities
Others
Download your FREE sample PDF copy of the Next-Generation Sequencing Services Market today and explore key data and trends.
0 notes
Text
In Down syndrome mice, 40Hz light and sound improve cognition, neurogenesis, connectivity
New Post has been published on https://sunalei.org/news/in-down-syndrome-mice-40hz-light-and-sound-improve-cognition-neurogenesis-connectivity/
In Down syndrome mice, 40Hz light and sound improve cognition, neurogenesis, connectivity

Studies by a growing number of labs have identified neurological health benefits from exposing human volunteers or animal models to light, sound, and/or tactile stimulation at the brain’s “gamma” frequency rhythm of 40Hz. In the latest such research at The Picower Institute for Learning and Memory and Alana Down Syndrome Center at MIT, scientists found that 40Hz sensory stimulation improved cognition and circuit connectivity and encouraged the growth of new neurons in mice genetically engineered to model Down syndrome.
Li-Huei Tsai, Picower Professor at MIT and senior author of the new study in PLOS ONE, says that the results are encouraging, but also cautions that much more work is needed to test whether the method, called GENUS (for gamma entrainment using sensory stimulation), could provide clinical benefits for people with Down syndrome. Her lab has begun a small study with human volunteers at MIT.
“While this work, for the first time, shows beneficial effects of GENUS on Down syndrome using an imperfect mouse model, we need to be cautious, as there is not yet data showing whether this also works in humans,” says Tsai, who directs The Picower Institute and The Alana Center, and is a member of MIT’s Department of Brain and Cognitive Sciences faculty.
Still, she says, the newly published article adds evidence that GENUS can promote a broad-based, restorative, “homeostatic” health response in the brain amid a wide variety of pathologies. Most GENUS studies have addressed Alzheimer’s disease in humans or mice, but others have found benefits from the stimulation for conditions such as “chemo brain” and stroke.
Down syndrome benefits
In the study, the research team led by postdoc Md Rezaul Islam and Brennan Jackson PhD ’23 worked with the commonly used “Ts65Dn” Down syndrome mouse model. The model recapitulates key aspects of the disorder, although it does not exactly mirror the human condition, which is caused by carrying an extra copy of chromosome 21.
In the first set of experiments in the paper, the team shows that an hour a day of 40Hz light and sound exposure for three weeks was associated with significant improvements on three standard short-term memory tests — two involving distinguishing novelty from familiarity and one involving spatial navigation. Because these kinds of memory tasks involve a brain region called the hippocampus, the researchers looked at neural activity there and measured a significant increase in activity indicators among mice that received the GENUS stimulation versus those that did not.
To better understand how stimulated mice could show improved cognition, the researchers examined whether cells in the hippocampus changed how they express their genes. To do this, the team used a technique called single cell RNA sequencing, which provided a readout of how nearly 16,000 individual neurons and other cells transcribed their DNA into RNA, a key step in gene expression. Many of the genes whose expression varied most prominently in neurons between the mice that received stimulation and those that did not were directly related to forming and organizing neural circuit connections called synapses.
To confirm the significance of that finding, the researchers directly examined the hippocampus in stimulated and control mice. They found that in a critical subregion, the dentate gyrus, stimulated mice had significantly more synapses.
Diving deeper
The team not only examined gene expression across individual cells, but also analyzed those data to assess whether there were patterns of coordination across multiple genes. Indeed, they found several such “modules” of co-expression. Some of this evidence further substantiated the idea that 40Hz-stimulated mice made important improvements in synaptic connectivity, but another key finding highlighted a role for TCF4, a key regulator of gene transcription needed for generating new neurons, or “neurogenesis.”
The team’s analysis of genetic data suggested that TCF4 is underexpressed in Down syndrome mice, but the researchers saw improved TCF4 expression in GENUS-stimulated mice. When the researchers went to the lab bench to determine whether the mice also exhibited a difference in neurogenesis, they found direct evidence that stimulated mice exhibited more than unstimulated mice in the dentate gyrus. These increases in TCF4 expression and neurogenesis are only correlational, the researchers noted, but they hypothesize that the increase in new neurons likely helps explain at least some of the increase in new synapses and improved short-term memory function.
“The increased putative functional synapses in the dentate gyrus is likely related to the increased adult neurogenesis observed in the Down syndrome mice following GENUS treatment,” Islam says.
This study is the first to document that GENUS is associated with increased neurogenesis.
The analysis of gene expression modules also yielded other key insights. One is that a cluster of genes whose expression typically declines with normal aging, and in Alzheimer’s disease, remained at higher expression levels among mice who received 40Hz sensory stimulation.
And the researchers also found evidence that mice that received stimulation retained more cells in the hippocampus that express Reelin. Reelin-expressing neurons are especially vulnerable in Alzheimer’s disease, but expression of the protein is associated with cognitive resilience amid Alzheimer’s disease pathology, which Ts65Dn mice develop. About 90 percent of people with Down syndrome develop Alzheimer’s disease, typically after the age of 40.
“In this study, we found that GENUS enhances the percentage of Reln+ neurons in hippocampus of a mouse model of Down syndrome, suggesting that GENUS may promote cognitive resilience,” Islam says.
Taken together with other studies, Tsai and Islam say, the new results add evidence that GENUS helps to stimulate the brain at the cellular and molecular level to mount a homeostatic response to aberrations caused by disease pathology, be it neurodegeneration in Alzheimer’s, demyelination in chemo brain, or deficits of neurogenesis in Down syndrome.
But the authors also cautioned that the study had limits. Not only is the Ts65Dn model an imperfect reflection of human Down syndrome, but also the mice used were all male. Moreover, the cognitive tests in the study only measured short-term memory. And finally, while the study was novel for extensively examining gene expression in the hippocampus amid GENUS stimulation, it did not look at changes in other cognitively critical brain regions, such as the prefrontal cortex.
In addition to Jackson, Islam, and Tsai, the paper’s other authors are Maeesha Tasnim Naomi, Brooke Schatz, Noah Tan, Mitchell Murdock, Dong Shin Park, Daniela Rodrigues Amorim, Fred Jiang, S. Sebastian Pineda, Chinnakkaruppan Adaikkan, Vanesa Fernandez, Ute Geigenmuller, Rosalind Mott Firenze, Manolis Kellis, and Ed Boyden.
Funding for the study came from the Alana Down Syndrome Center at MIT and the Alana USA Foundation, the U.S. National Science Foundation, the La Caixa Banking Foundation, a European Molecular Biology Organization long-term postdoctoral fellowship, Barbara J. Weedon, Henry E. Singleton, and the Hubolow family.
0 notes
Text
Cell2Sentence: Understanding Single-Cell Biology With LLMs

C2S-Scale
Imagine asking a cell about its status, activities, or drug responses and getting a simple English response. Today, Yale University and Cell2Sentence-Scale (C2S-Scale) announce a set of open-source large language models trained to understand biology at the single-cell level.
C2S-Scale simplifies complex biological data into “cell sentences” by connecting biology and AI. Researchers may then ask questions about specific cells, such as “Is this cell cancerous?” or “How will this cell respond to Drug X?” and obtain short, biologically based natural language replies.
C2S-Scale may:
Accelerate medication discovery and development
Customise therapy for better outcomes.
Democratise research by open-sourcing.
Help researchers improve disease prevention, treatment, and understanding.
Due to C2S-Scale's extensive research on textual representations of cells and biological information, language-driven single-cell analysis employing massive language models offers exciting new applications.
Single-cell RNA sequencing
Each individual possesses trillions of cells that build organs, fight infections, or deliver oxygen. Even in the same tissue, cells vary. ScRNA-seq evaluates gene expression to determine cell function.
However, single-cell data are massive, complicated, and hard to assess. Gene expression measurements for each cell can be represented by thousands of values and analysed using specialised tools and models. Single-cell analysis is slow, hard to scale, and exclusively for experts.
Imagine translating hundreds of numbers into language words that language models and humans could understand. What if we could simply communicate with a cell about its feelings, actions, and reactions to treatment or illness? Understanding biological systems from cells to tissues might transform disease research, diagnosis, and treatment.
In the “Scaling Large Language Models for Next-Generation Single-Cell Analysis” session, Google is excited to introduce Cell2Sentence-Scale (C2S-Scale), a set of robust, open-source LLMs that “read” and “write” biological data at the single-cell level. We'll discuss single-cell biology, how cells become word sequences, and how C2S-Scale opens new biological research pathways.
From cells to sentences
C2S-Scale converts each cell's gene expression profile into a "cell sentence," a list of the cell's most active genes ranked by expression level. This lets natural language models like Google Gemini or Gemma be applied to scRNA-seq data.
Language-based interfaces improve single-cell data accessibility, interpretability, and flexibility for Google. Much of biology is described in text, such as gene names, cell types, and experimental information, so LLMs can analyse and understand this data.
The C2S-Scale model family
C2S-Scale extends Google's Gemma open model family for biological reasoning utilising data engineering and intelligently written prompts that include cell words, information, and other biological context. Since the core LLM architecture has not changed, C2S-Scale may leverage the large ecosystem, scalability, and infrastructure of general-purpose language models. LLMs trained on over 1 billion tokens from scientific literature, biological information, and real-world transcriptome datasets are the result.
The C2S-Scale series of models, with 410 million to 27 billion parameters, was built to meet scientific needs. Smaller models are easier to deploy and change with less computational power, making them ideal for exploratory investigations or low-resource environments. Larger models perform better on many biological processes but require more processing resources. By offering this range of model sizes, users may choose the model that best fits their use case while considering computation, speed, and performance. Every model will be open-source for development and use.
The C2S-Scale can accomplish what?
Biology chat: Single-cell data questions answered
Imagine someone asking, “How will this T cell react to anti-PD-1 therapy, a common cancer treatment?”
C2S-Scale models may answer in regular language using cellular data and biological knowledge from pre-training, as seen on the left. The right image shows how conversational analysis allows academics to interact with their data in a way that was previously impossible using natural language.
Use natural language to interpret data
Cell2Sentence-Scale can automatically summarise scRNA-seq data at various complexity levels, from cell type classification to tissue or experiment summaries. This helps researchers reliably and rapidly comprehend new datasets without writing complex algorithms.
Biological scaling laws
Google's major finding is that biological language models scale well and function consistently with model size. Larger C2S-Scale models perform better in biological tasks including producing cells and tissues and categorising cell kinds.
The parameter-efficient regime showed that model size consistently increased dataset interpretation semantic similarity scores. As the model reached 27 billion parameters, comprehensive fine-tuning greatly increased tissue gene overlap. This pattern is comparable to general-purpose LLMs and shows that biological LLMs will improve with additional data and computing, creating more powerful and generally applicable biological discovery tools.
Predicting cell fate
One noteworthy usage of Cell2Sentence-Scale is predicting a cell's reaction to a perturbation like a drug, gene deletion, or cytokine exposure. A baseline cell sentence and treatment description can be used to generate a gene expression sentence by the model.
This ability to reproduce biological activity in silico speeds drug discovery, personalised therapy, and prioritising investigations before they are done in the lab. C2S-Scale advances the production of realistic "virtual cells," which may replace conventional cell lines and animal models faster, cheaper, and more morally.
Reinforcement learning optimisation
As reinforcement learning refines huge language models like Gemini to follow instructions and respond in human-aligned ways, Google improves Cell2Sentence-Scale models for biological thinking. Applying semantic text evaluation reward functions like BERTScore trains C2S-Scale to deliver informative and physiologically correct responses that match the dataset. This guides the model towards scientifically useful responses in complicated tasks like therapeutic therapy simulation.
Try it
GitHub and HuggingFace now host Cell2Sentence resources and models. It encourages you to experiment with your own single-cell data, examine these technologies, and uncover the possibilities of teaching robots life language one cell at a time.
#technology#technews#govindhtech#news#technologynews#cells#Cell2Sentence#RNA sequencing#Single-cell#C2S-Scale#C2S
0 notes
Photo

Speaker confirmed for #XMeeting! 🚨 We are excited to welcome Professor Eduardo da Veiga Beltrame! An expert in developing methods and tools for analyzing single-cell omics data, Professor Beltrame aims to simplify biological data exploration and broaden access for researchers from all backgrounds. His research on single-cell RNA sequencing and interactive tools powered by machine learning is revolutionizing precision medicine! Don't miss the chance to learn from someone at the forefront of biological data science! 💻🧬 #XMeeting #SingleCell #RNAseq #Bioinformatics #PrecisionMedicine #DataScience#bioinformatica
0 notes
Text
Spatiotemporal Omics Market | BIS Research

For details, visit our page BIS Research
According to BIS Research, Spatiotemporal omics is an advanced field in life sciences that merges molecular profiling with spatial and temporal data to provide deeper biological insights. It enables researchers to study RNA, DNA, and proteins within their original tissue context, maintaining spatial organization often lost in traditional methods. This approach reveals how cells interact and change over time, offering valuable data for cancer research, neuroscience, and immunology. Clinically, it supports precision medicine by identifying biomarkers, refining diagnostics, and guiding targeted treatments. As adoption grows, spatiotemporal omics is transforming both research and clinical applications with unprecedented resolution and contextual understanding of biological systems.
The Spatio OMICS Market is expected to grow at a significant rate due to advancements in sequencing and imaging technologies, and expansion of research and development funding.
Key Market Drivers
Technological Advancements: Cutting-edge sequencing and imaging tools have enhanced the resolution and scalability of spatial omics, making them more practical for research and clinical use.
Rising Demand for Precision Medicine: Spatiotemporal omics enables personalized treatment strategies by uncovering spatially-resolved biomarkers, driving interest from pharmaceutical and biotech companies.
Increased R&D Investment: Government and institutional funding for genomics and precision medicine is accelerating the development and adoption of spatial omics technologies globally.
Market Challenges
High Technology Costs: The advanced instruments and reagents required for spatiotemporal omics are costly, making adoption challenging for many academic and smaller research institutions. This financial barrier limits access despite rising interest in spatial biology.
Complexity of Data Analysis: Spatiotemporal omics generate vast, high-dimensional datasets combining molecular and imaging data. Processing this information demands specialized software, computational infrastructure, and bioinformatics expertise. Without these, deriving actionable insights can be slow and resource-intensive.
Limited Skilled Workforce and Infrastructure: The field requires interdisciplinary skills in molecular biology, spatial imaging, and data science. However, a shortage of trained professionals and inadequate infrastructure in many regions slows down adoption and implementation across research and clinical environments.
To get detailed information on Spatiotemporal OMICS Industry, Click here!
Market Opportunities
Integration with AI and Advanced Analytics: The growing complexity of spatial omics data presents a significant opportunity for AI-driven solutions. Machine learning tools that enable automated image analysis, cell segmentation, and pattern recognition can greatly enhance data interpretation and scalability. Companies investing in intelligent analytics platforms are well-positioned to lead the market.
Development of Multi-Modal Omics Platforms: There is increasing demand for technologies that can analyze multiple molecular layers—such as DNA, RNA, and proteins—from the same tissue sample. Developing integrated multi-modal platforms offers a competitive edge, enabling researchers to gain more comprehensive biological insights from a single experiment.
Competitive Landscape and Regional Outlook
Key players in the spatiotemporal omics market include 10x Genomics, NanoString Technologies, Akoya Biosciences, Bruker Corporation, Vizgen, and RareCyte, all investing heavily in R&D, strategic acquisitions, and expanding product offerings.
Regionally, North America leads with early adoption, strong R&D support, and NIH funding. Europe is rapidly advancing, driven by research collaborations in countries like Germany and the U.K. Meanwhile, Asia-Pacific is emerging as a growth hub, with a focus on precision oncology and large-scale health studies.
Key Players
10x Genomics
NanoString Technologies
Akoya Biosciences
Bruker Corporation
Vizgen
RareCyte
Future Outlook
The spatiotemporal omics market is set for significant growth, driven by ongoing technological advancements and expanding applications across research and clinical fields. As spatial and temporal data integration improves, it will provide deeper insights into complex biological systems, leading to breakthroughs in disease understanding and treatment.
For a comprehensive analysis, refer to the full report by BIS Research: Spatiotemporal OMICS Market.
End Use Insights
This report offers actionable insights for organizations in the spatiotemporal omics market, providing strategies for product innovation, growth, and competition.
Innovation Strategy: It identifies opportunities for market entry and technology adoption, helping organizations stay ahead of the competition while meeting evolving customer demands.
Growth Strategy: The report outlines targeted growth strategies to optimize market share, enhance brand presence, and drive revenue expansion.
Competitive Strategy: It evaluates key competitors and offers practical guidance for maintaining a competitive edge in a rapidly evolving market.
Conclusion
The market for spatiotemporal omics is expected to increase significantly due to growing applications in clinical and research settings, growing need for precision medicine, and technical advancements. To keep a competitive edge, major competitors in the market are always improving their product offerings, investing in R&D, and inventing. Despite obstacles like exorbitant expenses and intricate data, the amalgamation of artificial intelligence and multi-modal platforms offers significant prospects. Organizations that use these insights can take advantage of development opportunities, overcome obstacles, and set themselves up for long-term success in the ever-changing spatiotemporal omics landscape.
0 notes
Text

Transcriptomics is the branch of molecular biology that focuses on the study of RNA transcripts produced by the genome under specific conditions. It provides insights into gene expression patterns, regulatory mechanisms, and cellular responses at a given time.
Key Techniques in Transcriptomics
RNA Sequencing (RNA-Seq) – A high-throughput method to analyze the complete transcriptome using next-generation sequencing (NGS).
Microarrays – A hybridization-based method that detects specific RNA sequences using complementary probes.
qRT-PCR (Quantitative Reverse Transcription PCR) – Used for precise quantification of specific mRNA levels.
Northern Blotting – A traditional method to detect specific RNA molecules.
Single-cell RNA-Seq (scRNA-Seq) – Studies transcriptomics at a single-cell resolution, helping to understand cellular heterogeneity.
Applications of Transcriptomics
Disease Biomarker Discovery – Identifying gene expression changes in diseases like cancer, diabetes, and neurodegenerative disorders.
Drug Development – Assessing how drugs influence gene expression at the cellular level.
Precision Medicine – Personalized treatment strategies based on an individual's transcriptomic profile.
Systems Biology – Understanding how genes interact in biological networks.
Functional Genomics – Linking transcriptome data with gene function and phenotype.
Challenges in Transcriptomics
Data Complexity – Large datasets require advanced bioinformatics tools for analysis.
RNA Stability – RNA is more prone to degradation than DNA, requiring careful handling.
High Cost – RNA-Seq, especially at single-cell resolution, remains expensive.
Biotechnology Scientist Awards
Visit Our Website : http://biotechnologyscientist.com
Contact Us : [email protected]
Nomination Link : https://biotechnologyscientist.com/member-submission/?ecategory=Membership&rcategory=Member…
#sciencefather#researchawards#Scientist#Scholar#Researcher #Transcriptomics #GeneExpression #RNASeq #SingleCellRNASeq #FunctionalGenomics #mRNA #tRNA #lncRNA #miRNA #Bioinformatics #NextGenSequencing #RNARegulation #Epitranscriptomics #qRTPCR #Microarrays #Omics #DifferentialExpression #CancerBiomarkers #StemCellResearch #RNAEditing #PrecisionMedicine #SystemsBiology #DrugDiscovery #ComputationalBiology #BiotechResearch
👉 Don’t forget to like, share, and subscribe for more exciting content!
Get Connected Here: =============
Facebook : https://www.facebook.com/profile.php?id=61572562140976
Twitter : https://x.com/DiyaLyra34020
Tumblr : https://www.tumblr.com/blog/biotechscientist
Blogger: https://www.blogger.com/u/1/blog/posts/3420909576767698629
Linked in : https://www.linkedin.com/in/biotechnology-scientist-117866349/
Pinterest : https://in.pinterest.com/biotechnologyscientist/
0 notes
Text
Fwd: Course: Online.IntroSingleCellAnalysis.Dec2-4
Begin forwarded message: > From: [email protected] > Subject: Course: Online.IntroSingleCellAnalysis.Dec2-4 > Date: 14 November 2024 at 05:12:20 GMT > To: [email protected] > > > > FINAL CALL - ONLINE COURSE – Introduction to Single Cell Analysis (ISCA01) > > https://ift.tt/aUCD1h8 > > 2nd - 4th December 2024 > > Please feel free to share! > > COURSE OVERVIEW -Take your RNA-Seq analysis to the next level with single > cell RNA-Seq. This technology allows insights with an unpredicted level of > detail, but that brings a new level of complexity to the data analysis. In > this course, we will learn about the most popular single cell platforms, > how to plan a scRNA-Seq experiment, deal with some of the many pitfalls > when analysing your data, and effectively gain exciting, and cell type > specific biological insights > > By the end of the course participants should: > > - Understand the basic principles of popular single cell platforms and > the pros and cons of the different technologies. > - Be able run standard software to process raw 10x Genomics and Parse > Bioscience data and interpret the outputs > - Understand how to use the ‘Trailmaker’ to quickly analyse > scRNA-Seq data. > - Understand the basics of the R Bioconductor ‘Seurat’ package, and > how to combine it with other tools. > - Understand how to perform appropriate data quality control and > filtering. > - Understand how to cluster cells both within and between samples, and > identify possible cell types of individual cells and clusters > - Understand how to use statistically robust methods to compare gene > expression between samples to identify cell type specific changes in > gene expression and potential pathways of interest. > > Please email [email protected] with any questions. > > Upcoming courses > > ONLINE COURSE – Introduction to Machine Learning using R and Rstudio > (IMLR02) This course will be delivered live > > ONLINE COURSE – Introduction to Single Cell Analysis (ISCA01) This > course will be delivered live > > ONLINE COURSE – Using Google Earth Engine in Ecological Studies (GEEE01) > This course will be delivered live > > ONLINE COURSE – Time Series Analysis and Forecasting using R and Rstudio > (TSAF01) This course will be delivered live > > ONLINE COURSE – Species Distribution Modelling With Bayesian Statistics > Using R (SDMB06) This course will be delivered live > > ONLINE COURSE – Remote sensing data analysis and coding in R for ecology > (RSDA01) This course will be delivered live > > ONLINE COURSE – Multivariate Analysis Of Ecological Communities Using > R With The VEGAN package (VGNR07) This course will be delivered live > > -- > > Oliver Hooker PhD. > > PR stats > > Oliver Hooker
0 notes
Text
"How to stay updated with the latest research in bioinformatics?
Bioinformatics is evolving rapidly, with new discoveries, algorithms, and datasets emerging every day. Whether you're a student, researcher, or professional, staying updated is crucial for growth in this interdisciplinary field. But with the overwhelming amount of information out there, how can you keep track of the latest research?
Here’s a step-by-step guide to staying informed about bioinformatics advancements:
📚 1. Follow Peer-Reviewed Journals
Leading journals publish cutting-edge bioinformatics research. Consider subscribing to:
Bioinformatics (Oxford Journals)
BMC Bioinformatics
Nucleic Acids Research (NAR)
Genome Biology
PLoS Computational Biology
Nature Biotechnology
📝 Pro Tip: Set up Google Scholar alerts for specific keywords like "machine learning in bioinformatics" or "single-cell RNA sequencing" to receive relevant papers directly in your inbox.
📰 2. Leverage Preprint Servers
Not all groundbreaking research is published in journals immediately. Many researchers upload their work to preprint servers:
bioRxiv (Preprints in biology and bioinformatics)
arXiv (Computational biology & AI in bioinformatics)
🧠 Why use preprints? They help you access fresh research before peer review, giving you a competitive edge.
💻 3. Follow Top Bioinformatics Blogs & Websites
Several platforms curate the latest developments in bioinformatics:
OMGenomics (Personal insights from bioinformatics professionals)
Bits of DNA (Exploring genomics and computational biology)
Biostars (Community-driven discussions on bioinformatics challenges)
SEQC Blog (Sequencing and bioinformatics trends)
🎙️ 4. Listen to Bioinformatics Podcasts
If you prefer learning on the go, podcasts are a great way to absorb new knowledge: 🎧 Best Bioinformatics Podcasts:
The Bioinformatics Chat
Genomics in 5 Minutes
The OmicsCast
🧑🤝🧑 5. Engage with the Bioinformatics Community
Joining discussions and interacting with experts helps you stay informed: 🔹 Reddit: r/bioinformatics, r/genomics 🔹 Twitter/X: Follow researchers and hashtags like #Bioinformatics, #ComputationalBiology 🔹 LinkedIn Groups: Bioinformatics Discussion Forum, AI in Bioinformatics 🔹 Slack & Discord: Join bioinformatics-specific communities for direct interaction
🎓 6. Take Online Courses & Webinars
Platforms like Coursera, edX, and BioPractify frequently update their courses to reflect the latest techniques in bioinformatics. Also, keep an eye out for:
Workshops by EMBL-EBI
Online tutorials from Galaxy Project & Bioconductor
📅 Tip: Many universities and conferences offer free webinars. Sign up for event notifications!
🔬 7. Attend Conferences & Hackathons
Networking at events helps you learn from researchers and industry leaders. Some key conferences include:
ISMB (Intelligent Systems for Molecular Biology)
RECOMB (Research in Computational Molecular Biology)
Genome Informatics Conference
BioHackathons (Hands-on experience with the latest tools)
🌍 Virtual & Hybrid Options: Many conferences now offer remote participation—take advantage of them!
🚀 8. Stay Hands-On with Open-Source Projects
Following GitHub repositories for bioinformatics tools and frameworks keeps you engaged with real-world applications. Some trending repositories:
Bioconductor (for R-based bioinformatics analysis)
Nextflow (for scalable data analysis workflows)
DeepVariant (AI-powered genome sequencing analysis by Google)
💡 Bonus: Contributing to open-source projects is a great way to learn while building your portfolio.
🔎 Final Thoughts
Bioinformatics is a dynamic field that blends biology, data science, and AI. Staying updated requires a multi-pronged approach—reading journals, engaging in online discussions, participating in hackathons, and continuously learning. By following these strategies, you’ll remain ahead in this ever-evolving domain.
📢 What’s your go-to method for staying updated in bioinformatics? Share your insights below! ⬇️✨
#Bioinformatics#ComputationalBiology#Genomics#MachineLearning#AIinBioinformatics#DataScience#BiotechCareers#BioinformaticsResearch#ScienceNews#Biostatistics#NextGenSequencing#BioinformaticsCommunity#ResearchTools#OpenSourceScience#GenomeAnalysis
1 note
·
View note