#tissue nucleic acid extraction kit
Text
https://globalarticlefinder.com/unlocking-forensic-and-medical-insights-with-advanced-body-fluid-dna-extraction/
Tissue and Body Fluid Nucleic Acid Extraction Kit
DNA extraction from body fluids is crucial in various fields, including forensic science, medical research, and clinical diagnostics. Over the years, significant advancements have been made in the techniques and methodologies employed for body fluid DNA extraction, enabling scientists and professionals to extract valuable genetic information from diverse bodily fluids. This article explores the recent developments in body fluid DNA extraction, using Tissue and Body Fluid Nucleic Acid Extraction Kit.
#nucleic acid purification kit#dna extraction kit#body fluid nucleic acid purification solution#nucleic acid extraction kit#dna purification kit#tissue nucleic acid extraction kit#body fluids extraction solution#testing solutions
1 note
·
View note
Note
Hello, Mr. Holmes! How are you?
So, long story short, I ended up with an optical microscope in my room more or less 4 months ago, with 200 previously made slides (secured in a proper box), and lots of new ones too, for me to prepare myself. I love microbiology (it's one of my hyperfixations, curse my neurodivergency) and now I love it even more (my mother has had to drag me away from the microscope - I named it Wesley - in the middle of the night multiple times now).
After much conversation, I finally convinced my mom to buy me the proper equipment to prepare the slides!
So, I'm sending this ask to you, as I know you also have a microscope and that you use it a lot: what kind of equipment do you recommend me buying (gloves, scalpel blades, tints, etc), while still remembering that all of the stuff needs to stay in my room (properly taken cared of by me, of course)?
For example, I'm unsure if different dyes are used for different smears and specimens due to it's affinity (I've noticed that on 'organic matter' slides, images are usually tinted purple or pink, while on plant-based slides, images are usually tinted green and blue, with a few red structures.) Considering that I don't have access to a mortuary, I will mostly make plant slides. There must be a difference in the dyes then, right?
Sorry for the long text! Hope this isn't too much of a bother.
- a 17-year-old :)
Congratulations on your new light microscope. I do hope you get the best out of it. I am overjoyed that someone else appreciates the art of microscopy and microbiology.
However, you need to be careful to not strain your eyes. It is recommended to take breaks every 15 minutes to close your eyes or focus on something in the distance to reaccommodate your eyes. And get up every 40 minutes, stretch and correct your posture. And it is recommended to not use a microscope more than 5 hours per day. John has to chase me away from my microscope sometimes to take a break when I sit there for hours, my posture like a Caridea.
Concerning equipment, you will obviously need a scalpel or other sharp blade to make very thin slices of your specimen, as thin as possible. And forceps to move your samples (best just get a whole dissection kit it has everything). Obviously slides and coverslips, pipettes for the stains or water, maybe some tubes. A pen to label your slides. In many staining procedures ethanol or acetone is also used. A waste jar to safely dispose of any chemicals, but be careful what you mix. A rack for staining and containers. I would recommend nitrile gloves, some people are sensitive to latex.
The dyes you use depend on the specimen. For example in histological slides of tissues hematoxylin and eosin are most commonly used (short HE-stain). That's what you most likely saw on your slides, it's blue, purple and pink.
Hematoxylin is a basic compound extracted and oxidised from the logwood tree (Haematoxylum campechianum), and it stains acidic compounds in the cells (or basophilic because they have an affinity for basic substances). For example nucleic acids like DNA or RNA get stained by hematoxylin because they are basophillic. And where are lots of nucleic acids? In the nucleus and ribosomes, that is why they appear blue to purple in the staining because they bind hematoxylin.
Eosin is an acidic compound, and stains basic or acidophilic compounds red or pinkish, like proteins, collagen, cytoplasm, extracellular matrix.

(Ductus epididymidis with HE-stain)
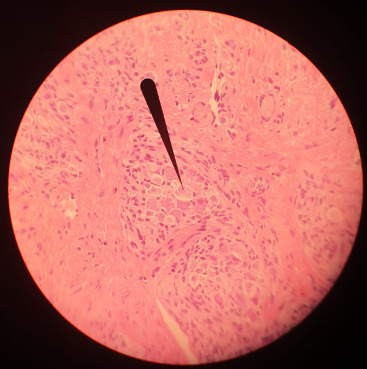
(Tongue HE-stain, pointer marking a ganglion; that is my picture)
Of course there are more specific stains for specific tissues like Golgi's silver staining for neurons.
For plants toluidine blue is often used, high affinity for acidic tissues, and can stain blue to green to purple.
It is often combined with safranin, a basic azine, which is probably the red stain you saw. It stains polysaccharides and lignin, woody parts of the plant. Safranin and astrablue is also often combined, astrablue stains non-lignified parts of the plant.
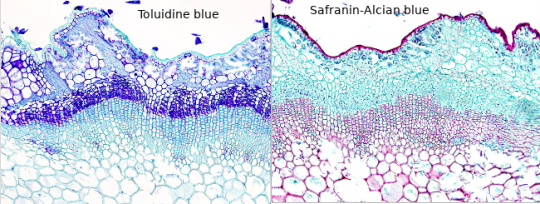
(Ulex europaeus stem; not my pictures I don't have any samples currently, source Atlas of plant and animal histology)
Safranin is also used in bacteriology, in the famous Gram staining. In Gram staining you use crystal violet (blue/purple), Lugol's iodine solution, then wash it with ethanol and add safranin (red) as a counter stain. Bacteria is gram-positive if the crystal violet stays in their thick murein cell wall, can't be washed out with the ethanol and the bacteria stays blue. Gram-negative appear red because of the counterstain.
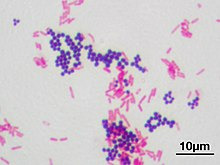
(Staphyloccocus aureus (violet, gram positive) & Escherichia coli (red, gram negative); not my picture, source Wikipedia)
However, I am not sure whether you have access to any of those substances, if they are too expensive for you or if they are too hazardous if used in your own room for a prolongued time. Of course those substances need to be stored properly, and your own room is probably not a good place, especially for ethanol or acetone. The fumes. I would recommend to ask your biology or chemistry teacher whether they can recommend anything further and where to buy said solutions in your area, and if they can't they are idiots. There are also many useful resources and tutorials on Youtube.
Another fascinating experiment for your microscope, that you can perform without buying any chemicals, is a hay infusion. You put hay into a container filled with water, and let it sit undisturbed for a week in a sunny area but not in direct harsh sunlight. During that time the microorganisms in the hay are reproducing in the solution, feeding on the polysaccharides of the hay. Protozoans also flourish in the hay infusion and eat the bacteria. It might get cloudy and a bit foul smelling (best not do it in your own room if you don't want to sleep next to a rotting smell).
When you put a drop of the solution onto a slide and look at it in the microscope, you should see a variety of microorganisms like bacteria (like Bacillus subtilis), amoeba, ciliates, heliozoa, algae et cetera. At different depths of the liquid you should find different kinds of organisms, because of differing oxygen content.
However, pathogens can also occur in the hay infusion so handle it carefully and work sterile, wash your hands properly.
And even if you don't work at a morgue you can still get tissue samples to experiment on, after all meat is sold in supermarkets, basically the same as a human body. And at the butchers they even sell organs like chicken hearts, pig kidney, liver, blood et cetera. Or observe your own hair under the microscope.
Which kind of samples and slides were included in your starter kit? Be careful to not leave them lying around in the sunlight, or the stain might fade. Always store them in the proper box.
#roleplay#rp#sherlock roleplay#sherlock rp#johnlock roleplay#johnlock rp#sherlock#bbc sherlock#sherlock holmes#sherlock holmes rp#sherlock holmes roleplay#science#scientist#research scientist#histology#microscope#microscopy#bacteria#bacteriology#pathology#anatomy#biology#chemistry#scientists#pictures#he stain#specimen#samples#slides#sherlock replies
47 notes
·
View notes
Text
DNA and RNA Sample Preparation Market is Estimated to Witness High Growth Owing to Increasing Adoption
The DNA and RNA sample preparation market involves processes associated with isolation, extraction, purification and quantification of nucleic acids DNA and RNA from various sources like tissues, blood, sperm, cells etc. for downstream applications in genomics, molecular diagnostics, personalized medicine and others. The sample preparation is a critical and initial step before conducting various genomic tests including Next Generation Sequencing, polymerase chain reaction and other assays. Growing awareness and adoption of precision medicine and genetic/molecular testing is driving demand for efficient nucleic acid isolation and downstream analysis.
The Global DNA and RNA Sample Preparation Market is estimated to be valued at US$ 2262.46 Mn in 2024 and is expected to exhibit a CAGR of 5.8% over the forecast period 2024 To 2031.
Key Takeaways
Key players operating in the DNA and RNA sample preparation are Agilent Technologies, Inc., Becton, Dickinson and Company, Bio-Rad Laboratories Inc., DiaSorin S.p.A, F. Hoffmann-La Roche, Miroculus, Inc., Illumina, Inc., PerkinElmer, Inc., QIAGEN, Sigma Aldrich Corp., Tecan Group AG, and Thermo Fisher Scientific, Inc. Growing prominence of personalized medicine is creating opportunities for development of new sample preparation methods and kits which can extract nucleic acids from various types of samples. Rising incidence of chronic and infectious diseases worldwide is increasing diagnostic testing which will propel sample preparation market growth. Global expansion of key market players through acquisitions and partnerships with regional diagnostic labs and research institutes will further augment market revenues.
Market Drivers
Increasing funding for Genomic and genetic research from government bodies as well as private sector is one of the key factors driving the DNA and RNA Sample Preparation Market Size. Government initiatives aimed at large scale population screening and clinical testing for various genetic disorders, infectious diseases and cancers are also creating demand for high throughput nucleic acid preparation. Growing geriatric population and rising healthcare spending in developing nations also provides growth opportunities for market players in the forecast period.
PEST Analysis
Political: Laws and regulations imposed by governments for research using DNA and RNA samples could impact the market. Changes in healthcare policies will also have effects.
Economic: Factors like GDP growth, income levels, healthcare spending will drive demand. Rise in research activities and focus on precision medicine boost the market.
Social: Growing awareness about personalized medicine and importance of genetic testing are important. Social trends also promote preventive healthcare and wellness.
Technological: Advancements in fields like next generation sequencing, lab automation, bioinformatics are key for market growth. Miniaturization and portability of equipment expand applications. Developments in sample collection and storage methods improve efficiency.
Geographical regions where the market in terms of value is concentrated include North America and Europe. North America accounts for the largest share in the global market due to presence of well-established healthcare industry and research institutes. Europe also captures notable share due to growing biotech sector and research funding.
The Asia Pacific region is projected to be the fastest growing market during the forecast period. This is attributed to factors such as increasing healthcare expenditure, growing awareness, expanding biotech industry and rising government investments in research. Countries like China, India offer growth opportunities as they focus on healthcare infrastructure development.
Get more insights on DNA And RNA Sample Preparation Market
Also read related article on Surgical Robots Market
Discover the Report for More Insights, Tailored to Your Language
French
German
Italian
Russian
Japanese
Chinese
Korean
Portuguese
Vaagisha brings over three years of expertise as a content editor in the market research domain. Originally a creative writer, she discovered her passion for editing, combining her flair for writing with a meticulous eye for detail. Her ability to craft and refine compelling content makes her an invaluable asset in delivering polished and engaging write-ups.
(LinkedIn: https://www.linkedin.com/in/vaagisha-singh-8080b91)

#Coherent Market Insights#DNA And RNA Sample Preparation Market#DNA And RNA Sample Preparation#RNA Sample Preparation#Nucleic Acid Extraction#Genetic Material Isolation#DNA Extraction#RNA Extraction#Molecular Biology#Genomic DNA
0 notes
Text
Biotechnology Reagents and Kits Market will grow at highest pace owing to increasing R&D activities in biopharmaceutical companies

Biotechnology reagents and kits are used for several diagnostic and research applications including isolation, purification, and analysis of DNA and RNA. Some key products in this market include cell and tissue culture reagents, chromatography reagents, electrophoresis reagents, immunohistochemistry reagents, nucleic acid extraction and purification reagents, PCR reagents, flow cytometry reagents, electrophoresis reagents, and separation and filtration reagents. Growing funding for biotechnology research along with rising prevalence of chronic diseases has increased the demand for biotechnology reagents and kits. Advancements in biologics and personalized medicine require reagents and kits for molecular analysis of patient samples which has fueled market growth.
The Global Biotechnology Reagents and Kits Market is estimated to be valued at US$ 718.8 Mn in 2024 and is expected to exhibit a CAGR of 23% over the forecast period 2023 to 2030.
Key Takeaways
Key players operating in the Biotechnology Reagents and Kits are Medtronic Plc, Hill-Rom Holdings, Inc. (Welch Allyn), iRythm Technologies, Inc., AliveCor, Inc., Vivalnk, Inc., Cardiac Insight Inc., VitalConnect, LifeSignals, Inc., Lâ€TMoreal Group, Dexcom, Inc., GENTAG, Inc., Abbott Laboratories, Koninklijke Philips N.V. and other prominent players. These players are focusing on new product development and launches to expand their product portfolio. For instance, in 2022 Medtronic Plc launched Clara Smart Pancreas System which uses RTSM technology to automatically suspend insulin delivery.
The demand for biotechnology reagents and kits is growing owing to increasing demand of personalized medicine, genomics applications and cell and gene therapy research. Pharmacogenomics, companion diagnostics and molecular biomarkers are driving the need for reagents and assays utilized in the discovery, development and commercialization of personalized medicine. Growing R&D spending on developing new biologics and rising application of big data analytics in drug development is also fueling market growth.
Technological advancements are expanding application of biotechnology reagents and kits in new areas. For example, next generation sequencing techniques have enabled large-scale sequencing of genomes which requires reagents for sample preparation and library preparation steps. Rapid diagnostics technologies are allowing point-of-care application of reagents for fast detection of infectious diseases. Automated instruments with integrated reagent kits are increasing efficiency and reproducibility of experiments in core labs.
Market trends:
1. Growing preference for personalized medicine/ precision medicine. Biomarker discovery and validation requires biomarker detection assays which is propelling demand for immunohistochemistry, PCR and other reagents.
2. Increasing focus on contract research and contract manufacturing. CROs and CMOs require a continuous supply of biotechnology reagents and kits for conducting research and manufacturing activities on behalf of biopharma companies.
Market Opportunities:
1. Emerging markets in Asia Pacific offer high growth potential. Countries like China, India and South Korea are witnessing increasing biotech research and bulk drug production which will drive the need for reagents in coming years.
2. Cell and gene therapy is a major area of focus. Reagents are essential for quality control testing, process development and scalable manufacturing of cell and gene therapies. This represents a lucrative opportunity for leading reagents players.
#Biotechnology Reagents and Kits Market Trend#Biotechnology Reagents and Kits Market Growth#Biotechnology Reagents and Kits Market Size
0 notes
Text
Nucleic Acid Isolation and Purification Market Future Trends to Look Out | Bis Research

Nucleic Acid Isolation and Purification is a fundamental molecular biology technique used to extract and purify nucleic acids (DNA or RNA) from various biological samples.
The global Nucleic Acid Isolation and Purification Market is projected to reach $5,615.9 million by 2033 from $2,922.8 million in 2023, growing at a CAGR of 6.75% during the forecast period 2023-2033.
Nucleic Acid Isolation and Purification Market Overview
Nucleic Acid Isolation and Purification is a critical process in molecular biology that involves extracting high-quality DNA or RNA from biological samples such as blood, tissue, cells, or microorganisms.
Key Steps in Nucleic Acid Isolation and Purification Market
Sample Collection and Preparation
Cell Lysis
Removal of Contaminants
Nucleic Acid Precipitation
Purification
Importance of Nucleic Acid Isolation and Purification
Nucleic Acid Isolation and Purification is a cornerstone technique in molecular biology and biotechnology, serving as a prerequisite for a multitude of critical applications.
Accurate Genetic Analysis
Genetic Engineering and Cloning
Clinical and Diagnostic Applications
Agricultural Biotechnology
Grab a look at the free sample page for more understanding click here !
Visit our Precision Medicine Vertical Page Click Here !
Market Segmentation
Product Type
Method Type
End Users
Applications
Region
Recent Developments in the Nucleic Acid Isolation and Purification Market
Qiagen N.V. introduced two groundbreaking additions to its sample technologies portfolio, i.e., the TissueLyser III that facilitates high-throughput disruption of diverse biological samples and the RNeasy PowerMax Soil Pro Kit that isolates high-purity RNA from challenging soil samples using advanced Inhibitor Removal Technology.
PerkinElmer introduced the CHEF Magnetic Bead Cleanup System, providing automated nucleic acid purification through advanced magnetic bead technology. This novel system would help automate the nucleic acid purification process efficiently.
Conclusion
In conclusion Nucleic Acid Isolation and Purification is a pivotal process in molecular biology, biotechnology, and medical diagnostics. Its significance lies in providing high-quality DNA and RNA essential for a wide range of applications, from basic research to clinical diagnostics, forensic science, and agricultural biotechnology.
The ability to isolate and purify nucleic acids effectively underpins many scientific and medical advancements, making it an indispensable technique in the pursuit of knowledge and the development of new technologies and therapies. As the field evolves, the refinement and expansion of these processes will continue to drive progress across multiple disciplines.
0 notes
Text
DNA Extraction Kits Market is Estimated to Witness High Growth Owing to Increasing Demand for Nucleic Acid-Based Molecular Tests

DNA extraction kits are essential tools used to isolate and purify DNA from a variety of biological samples in life science research and clinical diagnostics. DNA extraction kits can purify DNA from peripheral blood, tissue, cells, plasma, serum, saliva, stool, and other biological samples. Purified DNA is then utilized in various downstream applications including PCR, DNA sequencing, molecular testing, and synthetic biology. Common uses of DNA extraction kits in clinical applications include cancer diagnostics, forensic testing, prenatal testing, infectious disease testing, genetic testing, and personalized medicine.
The global DNA extraction kits market is estimated to be valued at US$1.2 Bn in 2023 and is expected to exhibit a CAGR of 5.9% over the forecast period 2023-2031, as highlighted in a new report published by Coherent Market Insights.
Market Dynamics
Increasing demand for nucleic acid-based molecular diagnostic tests is expected to drive growth of the DNA extraction kits market size over the forecast period. Molecular diagnostic testing is gaining immense popularity over traditional diagnostics due to its accuracy, high sensitivity, and specificity. Growing prevalence of cancer and infectious diseases is propelling demand for rapid and accurate molecular testing which require purified DNA obtained from extraction kits. According to WHO, cancer burden is expected to grow to 27.5 million new cancer cases and 16.3 million cancer deaths by 2040. Technological advancements in extraction methods allowing purification of DNA even from complex biological samples is another factor fueling the DNA extraction kits market growth. Automated liquid handling workstations integrated with magnetic bead-based DNA extraction techniques have simplified workflow and increased throughput of purification process.
SWOT Analysis
Strength: DNA extraction kits provide an easy and efficient way to isolate and purify DNA from various biological samples like blood, saliva, and tissues. These kits use optimized procedures and reagents to reliably extract high yields of pure DNA. They minimize hands-on time and produce consistent results. Some kits are automated and high-throughput, allowing labs to extract DNA from large numbers of samples simultaneously in an easy and standardized manner.
Weakness: DNA extraction kits rely on consumables like columns, plates and reagents that need regular replenishment, increasing long-term costs. Additionally, automation requires larger initial investments. The quality of extracted DNA can vary between kits depending on their purification chemistry and protocols.
Opportunity: Growth in genomic researchareas like precision medicine, cancer research and genetic testing is driving increased demand for DNA extraction. Rising focus on pharmacogenomics and companion diagnostics also presents commercialization opportunities. Point-of-care extraction devices and portable battery-operated systems allow decentralized testing and expand the addressable market.
Threats: Competition from in-house extraction methods developed by research institutes poses a challenge. Stringent regulatory approvals for diagnostic-grade kits increase barriers. Intellectual property disputes between manufacturers also affect commercialization strategies.
Key Takeaways
The global DNA extraction kits market is expected to witness high growth over the forecast period driven by increasing genomics funding and expanding application areas.
The market is dominated by North America currently due to extensive R&D spending and presence of leading life science companies. Asia Pacific is poised to be the fastest growing regional market due to growing genomics research in countries like China and India as well as increasing healthcare investments.
Key players operating in the DNA extraction kits market are Qiagen N.V., Thermo Fisher Scientific, Hoffmann-La Roche AG, Merck KGaA, Agilent Technologies, Bio-Rad Laboratories Inc., Illumina, Inc., Promega Corporation, LGC Ltd., and Takara Bio, Inc. Companies are focused on portfolio expansion through new product launches. For example, in 2021 Thermo Fisher launched the MagMAX Cellular RNA Isolation Kit designed for high-throughput extraction of total RNA from cultured cells.
Get more insights on this topic: https://www.newswirestats.com/dna-extraction-kits-market-size-and-outlook/ Explore more information, Please visit:https://masstamilan.in/savoring-life-the-essential-role-and-health-benefits-of-spices-in-everyday-cuisine/
#DNA Extraction Kits#DNA Extraction Kits Market#DNA Extraction Kits Market size#DNA Extraction Kits Market share#Coherent Market Insights
0 notes
Text
INDICAL BIOSCIENCE | Sample preparation | Online Shop
Indical sample preparation kits are designed to facilitate the processing and preparation of various types of animal samples for testing and analysis. These kits typically contain specific reagents and components necessary for sample collection, stabilization, extraction, and purification. They aim to streamline the sample preparation process, enhance the quality of extracted biomolecules, and improve the overall accuracy and reliability of veterinary diagnostic tests.
Here are some common types of veterinary sample preparation kits:
DNA/RNA Extraction Kits: These kits provide reagents and protocols for isolating DNA or RNA from different animal sample types, such as blood, tissue, swabs, or feces. They often include lysis buffers, spin columns, purification columns, and other necessary components to efficiently extract and purify nucleic acids for downstream molecular diagnostics, such as PCR-based tests or sequencing.
Protein Extraction Kits: These kits are designed to extract proteins from animal tissues, body fluids, or cells. They typically provide optimized buffers, inhibitors, and protease cocktails to ensure efficient protein extraction while preserving their integrity and activity. The extracted proteins can be used for various applications, including immunological assays, enzyme activity measurements, or protein profiling studies.
Sample Stabilization Kits: Sample stabilization is crucial to maintain the integrity of biomarkers or analytes in veterinary samples during collection, transport, and storage. Stabilization kits often contain specific reagents or collection tubes that prevent degradation or changes in the target analytes. These kits are particularly useful for veterinary diagnostics that require the analysis of unstable or time-sensitive analytes, such as hormones, metabolites, or certain pathogens.
Microbiological Sample Preparation Kits: These kits are designed to prepare samples for microbial analysis in veterinary diagnostics. They may include selective enrichment media, growth supplements, and inhibitors to promote the growth of target microorganisms while inhibiting the growth of unwanted contaminants. Microbiological sample preparation kits can be used for bacterial, viral, or fungal detection in various animal samples, such as swabs, fluids, or tissues.
It's important to note that the specific contents and protocols of veterinary sample preparation kits can vary depending on the manufacturer, target analytes, and sample types. Veterinary laboratories and clinics can choose kits tailored to their specific diagnostic needs and ensure compatibility with the subsequent assays or tests performed on the prepared samples.
Shop Now - https://shop.indical.com/en/sample-preparation/
0 notes
Text
Nucleic Acid Extraction System MultiEX 032

MultiEX 032 Automated Nucleic Acid Extractor can be used in Research Labs (Government & Academic Research), Commercial Labs (Pharmaceutical & BioTechnology Industries), and Molecular Diagnostics Labs.
It is an automated nucleic acid extraction and purification system which can process up to 32 samples per run. The system uses Magnetic Bead Separation technology and has the capability of extracting and purifying nucleic acids from a variety of biological samples, such as whole blood, viruses, tissues, plants, animals, fungi, bacteria, cultured cells, swabs, feces, blood cards, etc.
The nucleic acid extraction machine comes with a user-friendly touch screen interface allowing the user to set purification parameters easily. The automated nucleic acid extraction system integrates all the purification phases, including binding, mixing, washing, and elution, and delivers high-efficiency, high-quality purification. The system works with any prefilled magnetic bead-based reagent kit.
Features:
● Compact & Lightweight: Help save your limited laboratory bench space.
● Stable and Efficient: Automated processing avoids discrepancies and errors caused by manual operations and increases the efficiency
● Ease of Use: A 10-inch color touch screen and user-friendly interface lets you easily set purification parameters
● Contamination Control: Built-in UV disinfection lamp eliminates cross-contamination.
●Advanced Stirring Technology: Stirring speed up to 10,000 mm/min enables effective mixing of samples with high viscosity (blood, feces, etc.).
● Multi Heating Modules: Multiple heating modules with individual heating controls ensures temperature accuracy for lysis and elution.
● Safe and Reliable: The operation will automatically suspend once the safety door opens and only resumes after the safety door is shut, which provides better protection for users.
0 notes
Text
What are the common misunderstandings in the process of magnetic bead nucleic acid extraction?
The use of biological magnetic beads for nucleic acid extraction is still a relatively novel nucleic acid extraction method in China. Compared with the traditional chloroform isoamyl alcohol extraction method and spin column kit method, this method is still understood by many people. There are also some misunderstandings in the process of using magnetic beads to purify nucleic acids.
Misunderstanding 1: The more magnetic beads used, the better the extraction effect
Many teachers like to increase the amount of magnetic beads when the extraction effect is not good. They think that adding a little more magnetic beads can attract more nucleic acids. I have to say that this idea is not advisable.
The main feature of magnetic beads is that they can be dispersed in a liquid or separated from a liquid phase in a solid state under the action of an external magnetic field. For any reagent system, the ratio of magnetic beads to liquid should have a certain threshold, exceeding a certain ratio Excessive magnetic beads will lose their dispersion characteristics because they cannot be uniformly dispersed in the liquid, and the efficiency of contact between the nucleic acid magnetic beads and the liquid cannot be fully increased during the washing process. Excessive magnetic beads will also adsorb more impurities, which has a great influence on the effect of impurity removal. Even sometimes, too many magnetic beads will adsorb protease, lysozyme and other functional components that play a major role in the liquid system, resulting in low efficiency of the entire kit. In many cases, when the extraction effect is not good, reducing the amount of magnetic beads used is the best way to improve the extraction effect.
Normally, the amount of reference magnetic beads given by the magnetic bead method kit is slightly excessive. Therefore, it is not often necessary to increase the amount of magnetic beads to improve the adsorption efficiency. However, if it is determined that the extraction effect is not caused by insufficient amount of magnetic beads Well, it is possible to improve the extraction effect by increasing the amount of magnetic beads within a certain range.
Take GNT-02 series magnetic beads as an example. When extracting macro samples (plant tissue, whole blood, etc.), the usual dosage is 10ul/time; when extracting trace samples (such as serum free DNA, buccal swabs, etc.), magnetic beads The dosage is 15~20ul/time. If you need to exceed this usage, you need to communicate with a technical engineer.
...
Click to visit for details:https://www.aisenbio.com/news/common-misunderstandings-of-magnetic-bead-method-of-nucleic-acid-extraction/
0 notes
Text
Cell Isolation Market Trends, Revenue, Major Players, Share Analysis & Forecast Till 2026
According to the current analysis of Reports and Data, The global cell isolation market was valued at USD 4.9 billion in 2018 and is expected to reach USD 18.28 billion by the year 2026, at a CAGR of 17.7%. Cell isolation is the process of extracting a specialized cell from a heterogeneous mixture and then process it to identify its properties and replicate it to develop new therapies. For the determination of appropriate separation technique, an exhaustive analysis of the cell size, cell behavior, density, antigen status, and hydrophobic surface properties are done.
Cell isolation plays a very vital role in the diagnostics and research of chronic diseases. It helps in drug discovery by studying the behaviour of the cells and their response to disease and drugs. This technique of drug discovery helps to generate medicines that can be used for the treatment of various diseases such as cancer, genetic disorders, and autoimmune diseases. With incidents of chronic diseases on the rise across the world, the research, drug development, and clinical trials on various cell-based therapies also need to be increased. Therefore, the demand for cell isolation market will also have a boost. This will be a significant factor fuelling the growth of the cell isolation market. The cell separation techniques play a vital role in personalized medicines, which are used for early detection of disease, selection of appropriate treatment, and determining the prognosis of the therapy. All these factors have contributed towards a positive dynamic growth curve of this market, and it is expected to keep growing in the coming years.
Get a sample of the report @ https://www.reportsanddata.com/sample-enquiry-form/2237
The report provides an in-depth analysis of the key developments and innovations of the market such as research and development advancements, product launches, mergers & acquisitions, joint ventures, partnerships, government deals, and collaborations. The report offers a comprehensive assessment of key players in the market along with their global position, financial standing, business expansion plans, production and manufacturing capacity, and strategic alliances.
Key players in the market include
Thermo Fisher Scientific, Beckman Coulter, Becton, Dickinson and Company, GE Healthcare, Merck KgaA, Miltenyi Biotec, pluriSelect, STEMCELL Technologies Inc., Terumo BCT and Bio-Rad Laboratories Inc
Furthermore, the report segments Cell Isolation market on the basis of key product types and applications and provides details about the revenue growth, revenue CAGR, and revenue share each segment is expected to register over the forecast period.
Product (Revenue in USD Billion, 2018 - 2026)
Reagents, kits, media, and sera
Beads
Disposables
Centrifuges
Flow cytometers
Filtration systems
Magnetic-activated cell separator systems
Cell Type (Revenue in USD Billion, 2018 - 2026)
Differentiated Cells
Stem Cells
Technique (Revenue in USD Billion, 2018 - 2026)
Centrifugation
Surface marker
Filtration
Application (Revenue in USD Billion, 2018 - 2026)
Biomolecule Isolation
Cancer Research
Stem Cell Research
Tissue Regeneration & Regenerative Medicine
Vitro Diagnostics
End Use (Revenue in USD Billion, 2018 - 2026)
Research laboratories and institutes
Biotechnology and biopharmaceutical companies
Cell banks
Hospitals and diagnostic laboratories
For a better understanding of the global Cell Isolation market dynamics, a regional analysis of the market across key geographical areas is offered in the report. The market is spread across key geographical regions such as North America, Europe, Latin America, Asia Pacific, and Middle East & Africa. Each region is analysed on the basis of the market scenario in the major countries of the regions to provide a deeper understanding of the market. It provides insights into production and consumption patterns, supply and demand ratio, export/import, current and emerging trends and demands, market share, market size, revenue contribution, and presence of key players in each region.
Request a discount on the report @ https://www.reportsanddata.com/discount-enquiry-form/2237
In-depth regional analysis includes:
· North America (U.S., Canada, Mexico)
· Europe (Italy, U.K., Germany, France, Rest of EU)
· Asia Pacific (India, China, Japan, South Korea, Australia, Rest of APAC)
· Latin America (Chile, Brazil, Argentina, Peru, Rest of Latin America)
· Middle East & Africa (Saudi Arabia, U.A.E., South Africa, Rest of MEA)
To know more about the report @ https://www.reportsanddata.com/report-detail/cell-isolation-market
Benefits of the Global Cell Isolation Report:
• Comprehensive analysis of the opportunities and risks of the Cell Isolation market
• Detailed study of the latest product and technological developments and innovations of the Cell Isolation market
• Business strategies and plans are analysed for understanding the Cell Isolation market scenario
• Revenue forecast of Cell Isolation market for the forecast period 2020-2026
• Comprehensive analysis of the drivers, constraints, limitations, challenges, and opportunities
• Latest and emerging market trends analysis and their impact on product and application demand
• Study of recent M&A, joint ventures, collaborations, partnerships, product launches and brand promotions among others
• Extensive SWOT analysis and Porter’s Five Forces analysis along with investment return analysis and feasibility
Request a customization of the report @ https://www.reportsanddata.com/request-customization-form/2237
Thank you for reading our report. Please get in touch with us to know more about the report and customization of the report. Our team will ensure the report is tailored to meet your requirements.
Browse More Reports :
ELISpot and Fluorospot Assay Market Research
Nucleic Acid Isolation and Purification Market Growth Rate
0 notes
Text
Nucleic Acid Testing Market Research Report
Global Nucleic Acid Testing Market
The GMI Research forecasts that the Nucleic Acid Testing Market is witnessing an upsurge in demand over the forecast period. This is mainly due to the rise in demand for PCR kits and technological advancement across research & health care industries.
Request for a FREE Sample Report on Nucleic Acid Testing Market
Introduction of the Nucleic Acid Testing Market:
A nucleic acid test (NAT), also called nucleic acid amplification test (NAAT), is a pioneering technique used to detect a specific nucleic acid, virus, or bacteria that reacts as a disease-causing pathogen from biological samples such as blood, tissues, urine, etc. The three significant steps involved in the test are sample collection and nucleic acid (NA) extraction, target NA amplification, and target amplicon NA detection. In addition, the test includes an amplification step for the fallback of the genetic material.
Key Players of the Global Nucleic Acid Testing Market:
· Abbot Laboratories
· Danaher Corporation
· F. Hoffmann-La Roche Ltd.
· Thermo Fisher Scientific Inc.
· Merck KGaA
· Dickinson and Company
· Agilent Technologies
· Bio-Rad Laboratories Inc.
· Illumina, Inc.
· Qiagen
· Becton
Nucleic Acid Testing Market Dynamics (including market size, share, trends, forecast, growth, forecast, and industry analysis)
Several prominent drivers proliferating the growth of the global nucleic acid testing market include the introduction of novel and progressive NAT testing platforms, augmenting demand of kits from blood transfusion centers and the blood banks, along with ongoing technological advancements. For example, a team of researchers from the National University of Singapore (NUS) in September 2018 developed a moveable, user-friendly device for the quick and precise screening of diseases named envision. This device is designed to apprehend an extensive range of diseases, including transpiring infectious diseases such as Zika, Ebola, and others. In addition, the surging consciousness and rising molecular diagnosis, enhancing detection rate in developing countries, and switch from curative healthcare to preventive healthcare are some other factors responsible for the market growth. Moreover, the mounting occurrence of genetic diseases, cancer, and infectious diseases including hepatitis, dengue, and malaria, are projected to strengthen the nucleic acid testing market size. However, the requirement of dedicated infrastructure facility, insufficient trained technical expertise, and high sensitivity are hindering the nucleic acid testing market share.
Nucleic Acid Testing Market Segmentation:
Segmentation by Product Type
· Nucleic Acid test Kits
o Transcription-mediated amplification (TMA)
o Polymerase chain reaction (PCR)
o Ligase chain reaction (LCR)
o Whole genome sequencing
· Consumables
Segmentation by Indication Revenue
· Infectious diseases
· Cancer
· Forensic Testing
· Others
Segmentation by End User Revenue
· Hospitals
· Pathology laboratories
· Research Institutes
· Clinics
Segmentation by Region:
· North America
o United States of America
o Canada
· Asia Pacific
o China
o Japan
o India
o Rest of APAC
· Europe
o United Kingdom
o Germany
o France
o Spain
o Rest of Europe
· RoW
o Brazil
o South Africa
o Saudi Arabia
o UAE
o Rest of the world (remaining countries of the LAMEA region)
About GMI Research
GMI Research is a market research and consulting company that offers business sights and market research reports for every enterprise, including small & medium enterprises and large organizations. Our research team helps the clients to understand the impact of market dynamics such as market size, share, drivers, growth opportunities, and other aspects. We have a team of analysts and industry experts who conduct market intelligence studies to ensure relevant and fact-based research across a wide range of sectors such as FMCG, Technology, Energy, Healthcare, and other industries. We collect relevant information about the industry using both internal and external databases. Our main focus is to keep our clients abridged of the emerging opportunities and challenges in a wide range of industries. We provide step-by-step assistance to our client through strategic and consulting services to reach a managerial and actionable decision. Featured in the ‘Top 20 Most Promising Market Research Consultants’ list of Silicon India Magazine in 2018, we at GMI Research are always looking forward to helping businesses stay ahead of the curve.
Media Contact
Company Name: GMI RESEARCH
Contact Person: Sarah Nash
Email: [email protected]
Phone: Europe – +353 1 442 8820; US – +1 860 881 2270
Address: Dublin, Ireland
Website: www.gmiresearch.com
0 notes
Text
https://www.pressreleasepost.com/unlocking-forensic-and-medical-insights-with-advanced-body-fluid-dna-extraction/
Tissue and Body Fluid Nucleic Acid Extraction Kit
DNA extraction from body fluids is crucial in various fields, including forensic science, medical research, and clinical diagnostics. Over the years, significant advancements have been made in the techniques and methodologies employed for body fluid DNA extraction, enabling scientists and professionals to extract valuable genetic information from diverse bodily fluids. This article explores the recent developments in body fluid DNA extraction, using Tissue and Body Fluid Nucleic Acid Extraction Kit.
#nucleic acid purification kit#dna purification kit#body fluid nucleic acid purification solution#nucleic acid extraction kit#dna extraction kit#tissue nucleic acid extraction kit#testing solutions#body fluids extraction solution
1 note
·
View note
Text
How does the "universal" PCR laboratory carry out inspection projects?
PCR laboratory is also called gene amplification laboratory. Real-time fluorescent quantitative PCR technology is a leap forward in DNA quantitative technology. It is used to amplify specific DNA fragments, which can be regarded as special DNA replication in vitro. Through the DNA gene tracking system, the virus content in the patient's body can be quickly grasped with an accuracy of nanometer level. The real-time fluorescent quantitative PCR is the carrier to realize this technology. PCR experiments are often used to detect viral infectious diseases such as AIDS, hepatitis B, and poultry disease. By amplifying the genes contained in the virus, it is possible to detect whether some infected persons with low virus content contain specific viruses. The PCR Lab test method has the advantages of high sensitivity, high specificity, speed, and low sample requirements. Therefore, it is widely recognized by clinicians and has been widely used in clinical diagnosis in hospitals and diagnosis of poultry disease in various epidemic prevention and detection departments.
Driven by national policies and the development of the epidemic, hospitals at all levels in various regions have begun or are preparing to establish independent and compliant PCR laboratories factory, and they have become medical institutions with nucleic acid testing capabilities. However, due to the differences in the epidemic situation in various regions, some nucleic acid laboratories in long-term low-risk regions have almost stagnated except for emergency functions. This type of situation is also the most worrying issue for hospital leaders at all levels in setting up laboratories. They have invested money and energy but cannot bring income to the hospital.
The PCR laboratory is actually extremely powerful. Qualitative analysis and quantitative detection are its two biggest application directions. In addition to the new crown nucleic acid detection, what else can our "universal" PCR laboratory do?
Pathogen determination
The advent of PCR technology enables rapid and convenient pathogen detection. Because the false positive rate of PCR technology is too high, a positive result can be obtained as long as there is a small amount of pathogens, which cannot be used as a diagnostic basis, and it has clinical significance only when a certain number of pathogens exist. Therefore, it is particularly important to accurately quantify the template, and the results can be obtained quickly and accurately by using fluorescent technology PCR. PCR can be used to solve the "window period" problem of immunological testing, determine whether the disease is in a recessive or subclinical state, and when antibody testing cannot determine whether it is a current infection or a past infection.
Example: Novel coronavirus nucleic acid detection.
Methodology: fluorescence PCR method.
Uses: Used for in vitro qualitative detection of suspected cases of pneumonia, suspected clusters of patients with new coronavirus infection, and other patients who need to diagnose or identify new coronavirus infection.
Detection gene: new coronavirus (2019-nCoV) ORFlab and N gene, E gene.
Specimen types: upper respiratory tract specimens: throat swabs, nasal swabs; lower respiratory tract specimens: respiratory tract aspirates, alveolar lavage fluid samples, lung tissue biopsy specimens.
Detection process:
(1) Biosafety protection before nucleic acid testing (personal three-level protective wear by staff) → (2) Inactivation of samples (inactivation at 56°C for 30 minutes) → (3) Nucleic acid extraction of samples (manual, nucleic acid extractor) )→(4) PCR system preparation and template loading→(5) On-board testing and result analysis→(6) Disinfection after testing.
Other pathogen detection: HIV, hepatitis A, B and C, HPV, lung branches, lung coat, Epstein-Barr virus, adenovirus, cytovirus, A and B stream, Ebola virus, group B streptococcus.
Hospitals and departments suitable for development: children's hospitals, maternity and children's clinics, pediatric clinics, third parties, and hospitals of various levels in need.
Zhejiang KD Healthe Technology Co., Ltd was established in the year of 2015 and locates in the most beautiful city-Hangzhou, which is very near to Alibaba. We are the high-tech enterprise and focus on innovative health technology products. Since the outbreak of Corona virus, our factory has newly added seven production lines which were dedicated to the production of protective products, such as face mask, COVID-19 test kit, Infrared thermometer, safety glass, coverall and etc. We warmly welcome customers from all over the world.
0 notes
Text
Nucleic Acid Sample Preparation Market Future Trends to Look Out | Bis Research
Nucleic acid sample preparation refers to the set of processes and techniques employed to isolate, purify, and prepare nucleic acids (DNA or RNA) from various biological samples for downstream applications and analysis.
The global Nucleic Acid Sample Preparation Market is projected to reach $5,615.9 million by 2033 from $2,922.8 million in 2023, growing at a CAGR of 6.75% during the forecast period 2023-2033.
Nucleic Acid Sample Preparation Overview
This process is critical for ensuring the integrity and quality of nucleic acids, which are essential for reliable and accurate downstream applications such as polymerase chain reaction (PCR), sequencing, and gene expression analysis.
Key Steps in Nucleic Acid Sample Preparation
Sample Collection
Cell Lysis
Nucleic Acid Isolation
Purification
Quantification and Quality Assessment
Grab a look at the free sample page for more understanding click here !
Nucleic Acid Sample Preparation Important Considerations
Contamination - Avoiding contamination with nucleases, other nucleic acids, and environmental contaminants is critical. Use nuclease-free reagents and consumables.
Sample Handling - Gentle handling minimizes mechanical shearing of nucleic acids.
Control Samples - Always include control samples to monitor for contamination and process consistency.
Sample Collection and Preparation
Biological Source - Samples can be obtained from various sources such as blood, tissue, saliva, buccal swabs, or cultured cells.
Preservation - Immediate and appropriate preservation is crucial. For RNA, samples should be stabilized quickly using RNA stabilizing agents (e.g., RNAlater) to prevent degradation by RNases. For DNA, samples should be kept at low temperatures or preserved in ethanol.
Cell Lysis
Lysis Buffers: The choice of lysis buffer depends on the sample type and the nucleic acid to be extracted.
Mechanical Disruption: Methods like bead beating, sonication, or homogenization are often used to physically disrupt tough tissue samples.
Nucleic Acid Extraction
Phenol-Chloroform Extraction: This traditional method uses phenol and chloroform to separate proteins from nucleic acids, followed by ethanol precipitation to concentrate the nucleic acids.
Silica Column-Based Extraction: Common in commercial kits, this method involves binding nucleic acids to a silica membrane in the presence of chaotropic salts, followed by washing and elution.
Magnetic Beads: Nucleic acids bind to magnetic beads under specific conditions, allowing for easy washing and elution with the help of a magnetic field.
4. Purification
RNase and DNase Treatment: To remove contaminating RNA from DNA preparations, DNase treatment is applied, and vice versa for RNA preparations.
Column Wash Steps: Additional wash steps in column-based methods help to remove impurities such as proteins, salts, and other contaminants.
5. Quantification and Quality Assessment
Spectrophotometry: Measuring absorbance at 260 nm (A260) provides a quantitative estimate of nucleic acid concentration. The A260/A280 ratio indicates protein contamination, with ideal values around 1.8 for DNA and 2.0 for RNA.
Fluorometry: More sensitive than spectrophotometry, fluorometric assays (e.g., Qubit) use fluorescent dyes that bind specifically to nucleic acids.
Electrophoresis: Agarose gel electrophoresis can be used to assess the integrity and size distribution of the nucleic acids.
6. Storage
Short-Term Storage: Nucleic acids can be stored at 4°C for short-term use.
Long-Term Storage: For long-term preservation, DNA is stored at -20°C or -80°C, while RNA is best stored at -80°C to prevent degradation.
Nucleic Acid Sample Preparation Market Drivers
Increasing demand for molecular diagnostics
Advancements in Biotechnology and Genomics
Rising Research and Development Activities
Expansion of Biopharmaceutical Manufacturing
Key Market Players
Agilent Technologies, Inc.
Autogen, Inc.
Bio-Rad Laboratories, Inc.
Roche AG
Merck KGaA
and many others
Visit our Precision Medicine Vertical Page Click Here !
Market Segmentation
Product Type
End Users
Applications
Geography
Various different applications involved are as follows
Clinical Diagnostics
Biomedical Research
Pharmaceutical and Biotechnology Development
Agricultural Biotechnology
And many others
Recent Developments in the Nucleic Acid Sample Preparation Market Market
Qiagen N.V. introduced two groundbreaking additions to its sample technologies portfolio, i.e., the TissueLyser III that facilitates high-throughput disruption of diverse biological samples and the RNeasy PowerMax Soil Pro Kit that isolates high-purity RNA from challenging soil samples using advanced Inhibitor Removal Technology.
PerkinElmer introduced the CHEF Magnetic Bead Cleanup System, providing automated nucleic acid purification through advanced magnetic bead technology. This novel system would help automate the nucleic acid purification process efficiently.
Conclusion
In conclusion, the Nucleic Acid Sample Preparation market continues to experience significant growth and expansion driven by a multitude of factors. The increasing demand for molecular diagnostics, fueled by the prevalence of infectious diseases and genetic disorders, underscores the importance of nucleic acid-based testing in clinical settings.
Overall, the Nucleic Acid Sample Preparation market is poised for continued expansion, driven by ongoing advancements in technology, increasing research and development activities, and the growing demand for molecular diagnostics and personalized medicine.
0 notes
Text
New Post has been published on Abbkine - Antibodies, proteins, biochemicals, assay kits for life science research
How to choose appropriate NADP/NADPH Assay Kit?

NADP (coenzyme II) is the coenzyme of many redox reactions, including NADP+ (oxidation type) and NADPH (reduction type). NADP+ is also involved in biosynthesis reactions, such as lipid and nucleic acid synthesis. In animal cells, the oxidation stage of pentose phosphate pathway (PPP) is the main source of NADPH.
In this article, for NADP/NADPH Assay Kit, the author chose different brands (BioAssay Systems, Abcam, Biovision) and compared them with Abbkine CheKine™NADP/NADPH Assay Kit (Cat#: KTB1010) from several aspects such as assay principle, detection, sample type and size/price.
Summary:
These brands are the similar in Assay Principle, Detection and Sample type. Prices are different. The price of Abbkine NADP/NADPH Assay Kit (Cat#: KTB1010) is the lowest.
Besides good price, Abbkine NADP/NADPH Assay Kit (Cat#: KTB1010) has other advantages. The detection limit of Abbkine NADP/NADPH Assay Kit is 0.1 µM and linearity up to 4 µM NADP+/NADPH in 96-well plate assay.

Fig. Standard Curve of NADPH in 96-well plate assay. The y-axis is ΔOD and the x-axis is NADPH concentration (uM).
Details of different brands for NADP/NADPH Assay Kit are as follows,
BrandAssay PrincipleDetectionSample typeSize/PriceAbbkineThe assay is based on an enzymatic cycling reaction in which NADP+ is reduced to NADPH. NADPH reacts with a colorimetric probe that produces a colored product which can be measured at 565 nm. Detects NADP+, NADPH, or total NADP+/NADPH Tissue Extracts, Cell Lysate48T/$90
96T/$150BioAssay SystemsThe assay is based on an enzymatic cycling reaction in which NADP+ is reduced to NADPH. NADPH reacts with a colorimetric probe that produces a colored product which can be measured at 565 nm. Detects NADP+, NADPH, or total NADP+/NADPH Tissue Extracts, Cell Lysate100T/$379AbcamThe assay is based on an enzymatic cycling reaction in which NADP+ is reduced to NADPH. NADPH reacts with a colorimetric probe that produces a colored product which can be measured at 450 nm. Detects NADP+, NADPH, or total NADP+/NADPH Tissue Extracts, Cell Lysate100T/$615BiovisionThe assay is based on an enzymatic cycling reaction in which NADP+ is reduced to NADPH. NADPH reacts with a colorimetric probe that produces a colored product which can be measured at 450 nm. Detects NADP+, NADPH, or total NADP+/NADPH Tissue Extracts, Cell Lysate100T/$535
About Abbkine Scientific Co., Ltd.
Abbkine serves global scientists in the field of proteomics and cytology and is committed to the innovation and development of various scientific reagents related to proteomics and cytology, expecting to accelerate the pace of life science research and drug discovery. Proteomics products cover the preparation of samples (protein extraction, purification, coupling), protein quantification, antibodies and kits for protein detection. Cytology products involve cytokines (cell culture), cell status detection, cell staining, organelle extraction, cell metabolism and cytopathology reagents (kits). Abbkine relies on the product portfolio and unique marketing support as the main market strategy and product innovation mode, with ultimate aim to facilitate your research career.
0 notes
Text
Quantification of MicroRNAs for the Diagnostic Screening of Colon Cancer in Human Stool by Absolute Digital(d)PCR*| Lupine Publishers

Abstract
There is currently no validated micro(mi)RNA diagnostic stool test to screen for colon cancer (CC) on the market because of the complexity of fecal density, vulnerability of stool to daily changes, and the presence of three sources of miRNAs in stool (cell-free from fecal homogenates, exsosomal miRNAs from fecal exosomes, and fecal colonocytes). By employing earlier on a microarray miRNA experiment, using Affymetrix GeneChip miRNA 2.0 Arrays, on immunocaptured and enriched stool colonocytes of 15 subjects [three healthy controls and twelve colon cancer patients [three TNM stage 0-1 (e.g., polyps 1 cm, villous or tubvillous, or with high grade dysplasia), three stage 2, three stage 3, and three stage 4] in triplicates, this allowed for selection of a smaller panel of 14 preferentially expressed mature miRNAs associated with colon cancer (12 Up-Regulated, miR-19a, miR-20a, miR-21, miR-31, miR-34a, miR-96, miR-106a, miR-133a, miR-135b, miR-206, miR-224 and miR-302; and 2 Down-Regulated, miR-143 and miR-145). Then carrying out an absolute quantitative digital PCR on these 15 stool samples from TNM stages 0-4 on total small RNA extracted by immunocapture, followed by RT that employed a Custom TaqMan® miRNA Reverse Transcription (RT) Kit and TaqMan RT Primer Pool, and absolute quantification of miRNAs, in copies/μl, measured using a chip-based Absolute QuantStudio 3D Digital PCR analysis, allowed for validating the microarray results. To ensure that human and not bacterial small total RNA was chosen, coextraction protocols with E. coli K1 strain RS18 was carried out, followed by comparing Agilent electrophoretic patterns with human and bacterial electrophoretic patterns, and also random samples were sequenced using mRNA/miRNA sequencing, to ensure that human and not bacterial mRNA was chosen.
Introduction
Quantitative dPCR miRNA data presented in herein, show that the quantitative changes in the expression of a few mature miRNA genes in stool, which are associated with right and left colon cancer, would provide for a more convenient, sensitive and specific diagnostic screening molecular markers, more useful than markers currently available on the market, such as the low-sensitivity (<15%) fecal occult blood test(FOBT); result in better compliance; and is more economical than the invasive and expensive colon cancer colonoscopy exam, resulting in a higher probability of curing that cancer, if detected at the early TNM stages, and which becomes incurable and deadly if not diagnosed before metastasis.
Advantages of Using a MiRNA Diagnostic Colon Cancer Screening Test
The expression of individual genes may be altered by mutations in the DNA, or by a change in their regulation at the RNA or protein levels [1]. Epigenetic silencing is an important mechanism that contributes to gene inactivation in colorectal cancer (CRC) [2]. Analysis of promoter methylation of hypermethylated in cancer 1 (HIC1) gene in human stool showed it to be highly specific (98%) for both colon adenoma and carcinoma [3], but the sensitivity was quite low (31% for adenoma & 42% for all cancer), suggesting that an epigenetic marker only is not adequate for an accurate diagnostic screening, but a combination of genetic and epigenetic markers would be required to reliably identify CRC at an early disease stage [4]. Working with the stable DNA has been relatively easy compared to working with the fragile RNA molecule [1].A study by scientists at Exact Sciences Corp, Marlborough, MA, which markets a mutation-based DNA test “Cologuard”, assessed a newer version of a fecal DNA test for CRC screening using a vimentin methylation marker and another mutation DY marker plus nondegraded DNA in a limited sample of 44 CRC patients and 122 normal controls [5].
It cited a sensitivity of 88% and a specificity of 82%, only for advanced cancer, but not for the early adenoma stage. Besides, DNA mutation tests are not cost-effective, as screening for multiple mutations is expensive because these demanding mutation tests are not automated and are labor intensive. In addition, mutation detection in oncogenes and suppressor genes suffers from: a) the detection of mutations in these genes in fewer than half of large adenomas and carcinomas, b) the detection of gene mutations in non-neoplastic tissues, c) mutations found only in a portion of the tumor, and d) mutations often produce changes in the expression of many other genes [6,7]. Protein-based methods are currently not suited for screening and early diagnosis, either because proteins are not specific to one tumor or tissue type (e.g., CEA), their susceptibility to proteases, current lack of means to amplify proteins, no function is known for more than 75% of predicted proteins of multicellular organisms, there is not always a direct correlation between protein abundance and activity, and most importantly because detection of these markers exfoliately often signifies the presence of an advanced tumor stage. The dynamic range of protein expression in minimally-invasive body fluids (e.g., blood) is as large as 1010. Moreover, mRNA levels do not necessarily correlate with protein expressions.
Protein microarray studies revealed that protein expression vastly exceeds RNA levels, and only post translationally modified proteins are involved in signal transduction pathways leading to tumorigenesis. There is no well-documented protein test that has been shown in clinical trials to be a sensitive and a specific indicator of colon neoplasia, especially in early stages [8]. A serum proteomic study employing liquid chromatography (LC)-mass spectrometry (MS) carried out in a non-biased fashion failed to differentiate between individuals with large adenoma ( 1 cm) and normal individuals [9]. Compared to nucleic acids, proteomic research is a newer discipline; therefore, it will take considerable time to identify and validate proteins suitable for use as clinical markers, and resolve issues of bias and validations [10].On the other hand, a transcriptomic mRNA approach, has been shown to detect both adenomas and colon carcinomas with high sensitivity and specificity in preliminary studies [1], but no randomized, standardized, blinded prospective clinical studies have been carried out to validate the superiority of the mRNA approach.
A study indicated that a combination of a transcriptomic mRNA and miRNA expression signatures improves biomolecular classification of CRC [11]. Furthermore, not only does miRNAs regulate mRNA, but they also regulate protein expression. Two studies have shown that a single miRNA act as a rheostat to fine tune the expression of hundreds of proteins [12,13]. Hence, for CRC screening, miRNA markers are much more comprehensive and preferable to a DNA-, epigenetic-, mRNA- or a protein-based marker [14-18]. An added advantage for the use of the stable, nondegradable miRNAs by PCR expression, by chip-based methods, is its being automatable, making them much more economical and more easily acceptable by laboratory personnel performing these assays [4]. The discovery of small non-coding protein sequences,17-27 nucleotides long RNAs (microRNAs), has opened new opportunities for developing a non-invasive screening test for early diagnosis of many cancers. The latest miRbase release 22 on, March 12, 2018 [http://ww.mirbase.org] indicates the total number of miRNAs labeled “high confidence” has increased by 168, to 1996, than in the previous release [19].
MiRNA functions seem regulate development [20], apoptosis [21], and specific miRNAs are essential in oncogenesis [22,23], effective in classifying solid [24-26] and liquid tumors [27,28], and could serve as oncogenes or suppressor genes [29]. MiRNA genes are frequently found at fragile sites, as well as minimal regions of loss of heterozygosity, or amplification of common break-point regions [30], implying their involvement in carcinogenesis. MiRNAs have potential to serve as biomarkers for cancer diagnosis, prognosis and/or response to therapy [31,32]. Profiles of miRNA expression differ between normal and tumor tissues (33,34), suggesting that their expression profiles cluster similar tumor types together more accurately than expression profiles of protein-coding mRNA genes [33,34]. A study that examined global expression of 735 miRNAs in 315 samples of normal colonic mucosa, tubulovillus adenomas, adenocarcinomas proficient in DNA mismatch repair (pMMR), and defective in DNA mismatch repair (dMMR) representing sporadic and inherited CRC stages I-IV suggest involvement of common biologic pathways in pMMR and dMMR tumors in spite of the presence of numerous molecular differences between them, including differences at the miRNA level; indicating the need to pay attention to mismatch DNA repair (MMR) [34] .
Unlike screening for large numbers of messenger (m)RNA [1], a modest number of miRNAs is used to differentiate cancer from normal [35], and unlike mRNA, miRNAs in stool remain largely intact and stable for detection [36], therefore, leading to conclude that miRNA molecules are better markers to use for developing a reliable noninvasive diagnostic marker screen for colon cancer [14-18], since: a) the presence of the bacterium Escherichia coli does not hinder detection of miRNA by a sensitive technique such as dPCR [36], and b) the miRNA expression patterns are the same in primary tumor, or diseased tissue, as in stool samples [35-37]. The gold standard to which the miRNA test is compared to, has been “colonoscopy”, obtained from patients’ medical records [38]. However, because the low sensitivity guaiac FOBT is still the most commonly used screen in annual checkups (www.cancer.org) [39], this test should also be included for comparison with the proposed dPCR diagnostic miRNA screening approach in human stool.
Advantages of Stool Over Other Testing Media
Stool testing has several advantages over other colon cancer screening methods a s it is truly noninvasive and requires no unpleasant cathartic preparation, formal health care visits, or time away from work or routine activities [40-43]. Unlike sigmoidoscopy, it reflects the full length of the colorectum and samples can be taken in a way that represents the right and left side of the colon. It is also believed that colonocytes are released continuously and abundantly into the fecal stream, contrary to blood that is released intermittently as in guaiac fecal occult blood test (FOBT) [39]; therefore, this natural enrichment phenomenon partially obviates the need to use a laboratory-enrichment technique to enrich for tumorigenic colonocytes, as for example when blood is used for screen testing [44]. Furthermore, because testing can be performed on mail-in-specimens, geographic access stool screening is essentially unimpeded [4]. The American Cancer Society (ACS) has recognized stool-based molecular testing as a promising screening technology for CRC (www.cancer.org).
Isolation of colonocytes from stool, and comparing the Agilent electrophoretic (18S and 28S) patterns to those obtained from total RNA extracted from whole stool [45-47], and differential lysis of colonocytes by RT lysis buffer (Qiagen), could be construed as a validation that the electrophoretic pattern observed in stool (18S and 28S) is truly due to the presence of human colonocytes, and not due to stool contamination with Escherichia coli (16S and 23S). Taking into account that some exsosomal RNA will be released from purified colonocytes into stool, attempts must be made to correct for exsosomal RNA effect [48].
MiRNA dPCR Study Design
To test miRNAs as reliable, quantitative, sensitive and specific diagnostic biomarkers, for early non-invasive screening of colon cancer, using absolute dPCR test, preliminary work must be validated in a study using a nested case control epidemiology design and employing a prospective specimen collection, retrospective blind evaluation (Probe) of control subjects and test colon cancer patients, as specifically delineated by the National Cancer Institute’s Early Detection Research Network http://edrn.nci.nih.gov for cancer biomarker discovery studies. Selection of 14 miRNAs, 12 of them showed increased expression and 2 showed decreased expression for analysis of absolute miRNAs expression by a chipbased digital (d) PCR test is presented in Tables 1 & 2, and Figure 1. Absolute Quantitative Digital PCR Approach Digital PCR is a new approach to miRNAs quantification that offers an alternate method to qPCR for absolute quantification, by partitioning a sample of DNA or cDNA into many individual, parallel PCR reactions; some of these reactions contain the target molecule (positive), while others do not (negative).
Figure 1: Diagram illustrating QuantStudioTM 3D Digital PCR System Chip; ChipCase Lid (1); Digital PCR 20K 10 mm2 nanofluidic v2 chip (2), which contains 20,000 reaction wells; QuantStudioTM 3D Digital PCR Chip Case (3); Chip ID (4); Fill port (5); and Reaction wells, the 20,000 physical holes that suspend individual PCR reactions.
Table 1: Characteristics of Fourteen Up- or Down-Regulated MicroRNAs in Human Stool.
Table 2: Absolute Quantification of Up-/Down- Regulated miRNAs in Stool by QuantStudioTM 3D Chip-Based Digital PCR.
A single molecule can be amplified a million-fold or more. During amplification, TaqMan chemistry with dye-labeled probes is used to detect sequence-specific targets. When no target sequence is present, no signal accumulates. Following PCR analysis, the fraction of negative reactions is used to generate an absolute count of the number of target molecules in the sample, without the need for standards or endogenous controls. In conventional qPCR, the signal from wild-type sequences dominates and obscures the signal from rare sequences [49-51]. By minimizing the effect of competition between targets, dPCR overcomes the difficulties inherent to amplifying rare sequences and allows for sensitive & precise absolute quantification of the selected miRNAs. Applied Biosystem QuantStudio™ 3D instrument used in this research study, only performs the imaging and primary analysis of the digital chips. The chips themselves must be cycled offline on a Dual Flat Block GeneAmp® 9700 PCR System. or the ProFlex™ 2x Flat PCR System. The QuantStudio™ 3D Digital PCR System can read the digital chip in less than 1 minute, following thermal cycling [45].
It allows for one sample per chip; although, duplexing allows for analsis of two targets per chip. Sample prep for digital PCR is no different than for real-time qPCR, when using the QuantStudio™ 3D Digital PCR System. To figure out the concentration of cDNA stock from results, if one includes all of the necessary dilution factors into the AnalysisSuite™ software, the software will give the copies/μL in the stock. There are 2 dilutions that one needs to take into account:
a) The first is the dilution of the sample in the reaction,. and
b) The second is the dilution of the stock that one makes before adding it to the digital PCR reaction.
For example, if one wants to add 1 μL of a sample that has been diluted 1:10 from the stock. Thus, if one adds 1 μL of his/her sample to a 16 μL (final volume) reaction, the dilution factor of the sample is 1:16 or 1/16 = 0.0625. Since the stock has also been diluted 1:10 (0.1), one also need to factor this in. The final dilution factor to enter into the software is 0.0625 x 0.1 = 0.00625 (1:160). One can use either annotation to indicate the dilution factor in the AnalysisSuite™ software. If one enters that value into the “Dilution” column, the software will give the copies/μL in the starting material (stock).
The Poisson Plus algorithm for projects that contain QuantStudio™ 3D Chips with target, quantities >2000 copies/μL. The Poisson Plus algorithm corrects for well-to-well load volume variation, on a per Chip basis. This becomes important at higher target concentrations. There is also an option to export the Chip data as XML on the Export tab-thousands of discrete subunits prior to amplification by PCR, each ideally containing either zero or one (or at most, a few) template molecules [50]. Each partition behaves as an individual PCR reactions –as with real-time PCR-fluorescent FAM probes [or others, as VIC fluorescence]. Samples containing amplified products are considered positive (1, fluorescent), and those without product –with little or no fluorescence (i.e., are negative, 0). The ratio of positives to negatives in each sample is the basis of amplification. Unlike real-time qPCR, dPCR does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample, but it relies on Poisson Statistics to determine the absolute template quantity.
The unique sample partitioning step of dPCR, coupled with Poisson Statistics allows for higher precision than both traditional end point PCR. and qPCR methods; thereby allowing for analysis of rare miRNA targets quantitatively and accuratley [47,48]. The use of a nanofluidic chip, shown below, provides a convenient and straight forward mechanism to run thousands of PCR reactions in parallel. Each well is loaded with a mixture of sample, master mix, and Applied Biosystems TaqMan Assay reagents, and individually analyzed to detect the presence (positive) or absence (negative) of an endpoint signal. To account for wells that may have received more than one molecule of the target sequence, a correction factor is applied using the Poisson model. It features a filter set that is optimized for the FAM™, VIC®, and ROX™ dyes, available from Life Technologies [46]. The chips themselves must be cycled offline on a Dual Flat Block GeneAmp® 9700 PCR System. or the ProFlex™ 2x Flat PCR System. The QuantStudio™ 3D Digital PCR System can read the digital chip in less than 1 minute, following thermal cycling [45].
It allows for one sample per chip; although, duplexing allows for analsis of two targets per chip. Sample prep for digital PCR is no different than for real-time PCR, when using the Quant Studio™ 3D Digital PCR System. To figure out the concentration of cDNA stock from results, if one includes all of the necessary dilution factors into the AnalysisSuite™ software, the software will give the copies/sL in the stock. There are 2 dilutions that one needs to take into account:
a) The first is the dilution of the sample in the reaction,. and
b) The second is the dilution of the stock that one makes before adding it to the digital PCR reaction.
For example, if one wants to add 1 μL of a sample that has been diluted 1:10 from the stock. Thus, if one adds 1 μL of his/her sample to a 16 μL (final volume) reaction, the dilution factor of the sample is 1:16 or 1/16 = 0.0625. Since the stock has also been diluted 1:10 (0.1), one also need to factor this in. The final dilution factor to enter into the software is 0.0625 x 0.1 = 0.00625 (1:160). One can use either annotation to indicate the dilution factor in the AnalysisSuite™ software.
If one enters that value into the “Dilution” column, the software will give the copies/μL in the starting material (stock). The Poisson Plus algorithm for projects that contain Quant Studio™ 3D Chips with target, quantities >2000 copies/μL. The Poisson Plus algorithm corrects for well-to-well load volume variation, on a per Chip basis. This becomes important at higher target concentrations. There is also an option to export the Chip data as XML on the Export tab-thousands of discrete subunits prior to amplification by PCR, each ideally containing either zero or one (or at most, a few) template molecules [47]. Each partition behaves as an individual PCR reactions –as with real-time PCR—fluorescent FAM probes [or others, as VIC fluorescence]. Samples containing amplified products are considered positive (1, fluorescent), and those without product –with little or no fluorescence (i.e., are negative, 0). The ratio of positives to negatives in each sample is the basis of amplification. Unlike real-time qPCR, dPCR does not rely on the number of amplification cycles to determine the initial amount of template nucleic acid in each sample, but it relies on Poisson Statistics to determine the absolute template quantity.
The unique sample partitioning step of dPCR, coupled with Poisson Statistics allows for higher precision than both traditional and qPCR methods; thereby allowing for analysis of rare miRNA targets quantitatively and accuratley [47]. The use of a nanofluidic chip, shown below, provides a convenient and straight forward mechanism to run thousands of PCR reactions in parallel. Each well is loaded with a mixture of sample, master mix, and Applied Biosystems TaqMan Assay reagents, and individually analyzed to detect the presence (positive) or absence (negative) of an endpoint signal. To account for wells that may have received more than one molecule of the target sequence, a correction factor is applied using the Poisson model. It features a filter set that is optimized for the FAM™, VIC®, and ROX™ dyes, available from Life Technologies [46]. A workflow of the dPCR procedure by the QuantStudioTM 3D Digital PCR System is presented in (Figure 2) Workflow of a digital miRNAs PCR for colon cancer profiling in human colon tissue or stool samples.
Figure 2: Workflow of a digital miRNAs PCR for colon cancer profiling in human colon tissue or stool samples.
Digital PCR, however, has Several Tips to Follow:
i. A rough estimate of the concentration of miRNAs of interest has to be first carried out, in order to make appropriate dilutions, so that not too many partitions will get multiple copies that prevent accurate calculation of the copy number of miRNAs of interest;
ii. Non-template controls and a RT negative control must be set up for each miRNA, when using a “primer pool method” for retro-transcription;
iii. A chip-based dPCR method requires less pipetting steps, which reduces potential PCR contamination compared to another type of dPCR marketed by Bio-Rad Laboratories, thus called “Bio-Rad’s droplet digital PCR”, which requires multiple pipette transfers that potentially increase the risk of contamination [47], and
iv. Quant StudioTM 3D chip has 20,000 fixed reaction wells, whereas Bio-Rad’s droplet PCR relies upon the generation of droplets;
a step that could be extremely variable, as reported by Miotto et al (11,48) Absolute dPCR data tabulated in Tables 1 & 2, and presented graphically in Figure 1 below, which show 14 preferentially expressed mature miRNAs associated with colon cancer (12 Up-Regulated, miR-19a, miR-20a, miR-21, miR-31, miR- 34a, miR-96, miR-106a, miR-133a, miR-135b, miR-206, miR-224 and miR-302; and 2 Down-Regulated, miR-143 and miR-145) in stool samples from healthy controls, and stages 0-1 to 4 individuals with colon cancer . Standard deviations (sd) obtained from the one-way ANOVA, using the 5 level factor Type (normal, stage01, stage2, stage3, stage4) were calculated. The adjusted R-squared values representing the proportion of variation explained by Type are also reported. Type was statistically significant for every gene; all p-values were less than 0.000001 (no adjustments for multiple comparisons). These data are tabulated in Table 3 and shown graphically in Figure 1. For each gene on the graph in Figure 1, the min and max have been shown, in order to make the presentation clearer. At top left is high exxpression Value of 9985, which is the maximum value for that gene, at the bottom one finds the value for the minimum the colors range from dark blue (control) to orange (stage 4).
Table 3: Representation of SDs and R2 for miRNAs tested by absolute digital PCR.
The groups are also distinguished by line type: control (solid), stage 0-1 (long dash), stage 2 (dash), stage 3 (dot), stage 4 (dash nd dot). The figure is a parallel coordinate plot made in R, using the package MASS. For statistical analysis. Innovation and Clininical Significance of the dPCR-miRNA Diagnostic Stool Screening Approach Innovation lies in the collective use of many methods, such as: immunoparamagnetic beads to capture colonocytes from the harsh, noninvasive stool environment, whose extracted fragile total small RNA is stabilized shortly after stool excretion by commercial kits so it does not ever fragment, followed by standardized analytical quantitative miRNA dPCR-chip profiling in stool samples, which are neither labor intensive, nor require extensive sample preparation, to develop a panel of few stable miRNAs for absolute quantitative diagnostic screening of early sporadic colon cancer (stage 0-1), cheaper, with higher sensitivity and specificity than any other colon cancer screening test on the market . Isolation of colonocytes from stool samples is needed to provide a quantitative estimate of how our proposed miRNA method performs. Although we may miss exosome RNA, a parallel test could also be carried out on miRNAs obtained from stool samples to compare the extent of loss when colonocytes are only used, and an appropriate correction for exsosomal loss can be made [48]. Figure 3 Absolute Quantification of Up- or Down-Regulated miRNAs in Human Stool by Quant Studio TM 3D Digital PCR Chip System.
Figure 3: Absolute Quantification of Up- or Down-Regulated miRNAs in Human Stool by QuantStudioTM 3D Digital PCR Chip System.
Acknowledgments
We express our thanks to participating volunteers who provided stool samples for the study; Dr. Paul W. Vos of the Department of Biostatistics, East Carolina University for Statistical Analysis, and Dr. Clark D. Jeffries at Renaissance Computing Institute, University of North Carolina at Chapel Hill for his insight on bioinformatics’ analysis.
To know more about our Journal of Surgery click on https://lupinepublishers.com/surgery-case-studies-journal/
To know more about Lupine Publishers click on https://lupinepublishers.us/
To know more about Open access publishers click on Lupine Publishers
0 notes