#RNA-seq
Explore tagged Tumblr posts
Link
#Transcriptomic profile#Bladder cancer#Gene ontology#Gene enrichment#RNA-Seq#Differentially expressed genes
1 note
·
View note
Text
Unraveling Critical Insights: Designing New Gene Signature from RNA-seq Data
Creating a new gene signature from RNA sequencing (RNA-seq) data is a pivotal process in genomics. It has the potential to unlock critical insights into gene expression patterns. These patterns can further reveal biomarkers for various diseases, identify therapeutic targets, and elucidate underlying biological mechanisms governing health and diseases. Analyzing RNA-seq data enables researchers to discover genes that are differentially expressed in specific conditions, and provides a molecular signature that can be used to diagnose diseases, predict patient outcomes, and tailor treatments for individual patients.
Gene signatures are particularly valuable in personalized medicine as they can facilitate the development of targeted therapies based on the specific genetic makeup of a patient’s disease. In oncology, for instance, gene signatures can distinguish between different cancer subtypes, allowing for precise and effective treatment strategies. Gene signature applications extend to a wide range of diseases including cancer, cardiovascular disorders, neurological conditions, and infectious diseases.
This blog delineates the detailed process of developing a gene signature from RNA-seq data. It traces fundamental concepts like understanding RNA-seq technology and the significance of gene signatures. Further, it explores the preparation of data, including sample collection, data acquisition, and quality control followed by the processing and normalization of RNA-seq data, identification of differentially expressed genes (DEGs), and designing a robust gene signature. Eventually, it studies ways to apply and interpret these gene signatures in real-world scenarios, ensuring that the insights gained are actionable and relevant to ongoing research or clinical applications. Original Publish
0 notes
Note
oh dear. it seems that i am a -quickly looks at bio- teeny tiny protozoan unknown to science i hope a beautiful woman studies me and learns my teeny tiny protozoan secrets.
Damn I hope there's a gorgeous protozoa researcher out there to fulfill those wishes bc every model organism I've worked with has been in animalia
#quick list of what ive worked with#various passerine birds for field ecology volunteering#mice for neurophysiology research and as support staff#human cell lines for developing some rna seq methods#c elegans for a specific developmental process (current)#and three super secret specific model organisms i worked with during my phd rotations that would 100% doxx me
71 notes
·
View notes
Text
The Growing Frontier: An In-Depth Look at the Single-Cell Analysis Market
The Single-Cell Analysis Market was valued at USD 3.3 billion in 2023 and will surpass USD 8.7 billion by 2030; growing at a CAGR of 14.8% during 2024 - 2030. The field of life sciences is undergoing a transformative phase, with single-cell analysis emerging as a pivotal technique in modern biology and medicine. Unlike traditional bulk analysis methods that examine averages across populations of cells, single-cell analysis delves into the unique characteristics of individual cells, offering unprecedented insights into cellular diversity, disease mechanisms, and therapeutic targets. This article explores the current landscape, key drivers, and future prospects of the single-cell analysis market.
Single-cell analysis allows researchers to investigate the heterogeneity within a population of cells, which is crucial for understanding complex biological processes such as cancer progression, immune responses, and developmental biology. By examining individual cells, scientists can identify rare cell types, understand cell-to-cell variations, and gain insights into the dynamics of cellular networks. This level of detail is especially important in fields like oncology, immunology, and neurology, where subtle differences between cells can have significant implications for disease progression and treatment outcomes. The single-cell analysis market has experienced rapid growth over the past decade, driven by advancements in technology, increased research funding, and the growing recognition of the importance of cellular heterogeneity in biology and medicine.
Get a Sample Report: https://intentmarketresearch.com/request-sample/single-cell-analysis-market-3651.html
Key Drivers of Market Growth
Technological Advancements: Innovations in single-cell sequencing, microfluidics, and imaging technologies have significantly enhanced the accuracy, efficiency, and scalability of single-cell analysis. These advancements have made it easier for researchers to isolate, process, and analyze individual cells, driving adoption across various applications.
Rising Demand in Oncology: Cancer research is one of the primary areas driving the demand for single-cell analysis. The ability to identify and characterize rare cancer stem cells, understand tumor heterogeneity, and monitor the immune landscape of tumors has made single-cell analysis an indispensable tool in oncology.
Increased Funding and Collaborations: Governments, academic institutions, and private companies are increasingly investing in single-cell analysis research. Collaborative efforts between industry and academia are accelerating the development of new tools and applications, further fueling market growth.
Expansion of Applications: Beyond oncology, single-cell analysis is finding applications in immunology, neuroscience, stem cell research, and drug discovery. The versatility of this technology is broadening its appeal across multiple disciplines.
Challenges and Considerations
Despite its promising growth, the single-cell analysis market faces several challenges. The high cost of instruments and reagents remains a significant barrier for many research labs. Additionally, the complexity of data generated by single-cell analysis requires advanced bioinformatics tools and expertise, which can limit its accessibility to a broader range of researchers.
Furthermore, the standardization of protocols and data analysis methods is still evolving. Variability in sample preparation, sequencing techniques, and data interpretation can lead to inconsistencies in results, which is a critical issue that needs to be addressed to ensure the reliability of single-cell studies.
Get an insights of Customization: https://intentmarketresearch.com/ask-for-customization/single-cell-analysis-market-3651.html
Future Prospects
The future of the single-cell analysis market looks promising, with continued innovation and expansion into new research areas. Advances in artificial intelligence (AI) and machine learning (ML) are expected to play a significant role in improving data analysis and interpretation, making it easier for researchers to extract meaningful insights from complex datasets.
Moreover, as the cost of technology decreases and standardization improves, single-cell analysis is likely to become more accessible to a wider range of researchers, including those in smaller labs and developing countries. The integration of single-cell analysis with other omics technologies, such as proteomics and metabolomics, is also expected to open new avenues for research and personalized medicine.
Conclusion
The single-cell analysis market is at the forefront of a new era in biological research. As technology continues to advance and new applications emerge, this market is poised for substantial growth. The ability to study individual cells in detail is revolutionizing our understanding of health and disease, paving the way for more precise diagnostics, targeted therapies, and personalized medicine. For researchers, clinicians, and investors alike, the single-cell analysis market represents a dynamic and rapidly evolving frontier with significant potential to transform the life sciences landscape.
#Single-cell sequencing#Single-cell profiling#Single-cell RNA sequencing (scRNA-seq)#Single-cell genomics#Single-cell assay#Single-cell analytics
0 notes
Text
Rna-Seq Revelations: Pathway Analysis As The Genomic Compass
The advent of RNA-Seq technology has ushered in a new era of exploration, unraveling the mysteries encoded within our genes. Scientists delve into the vast sea of RNA data, and Pathway Analysis emerges as a powerful tool in their research. Let's explore a journey through the genomic landscape, where RNA-Seq becomes the compass guiding us through the intricacies of cellular pathways.
Read More :- https://www.myvipon.com/post/865752/Pathway-Analysis-The-Genomic-Compass-amazon-coupons
0 notes
Text
Reference saved in our archive
An interesting pilot study showing a probable biomarker for long covid.
Abstract
Introduction: Long COVID is a debilitating condition that lasts for more than three months post-infection by SARS–CoV–2. On average, one in ten individuals infected with SARS CoV- 2 develops Long COVID worldwide. A knowledge gap exists in our understanding of the mechanisms, genetic risk factors, and biomarkers that could be associated with Long COVID.
Methods: In this pilot study we used RNA-Seq to quantify the transcriptomes of peripheral blood mononuclear cells isolated from COVID-recovered individuals, seven with and seven without Long COVID symptoms (age- and sex-matched individuals), on average 6 months after infection.
Results: Seventy genes were identified as significantly up- or down-regulated in Long COVID samples, and the vast majority were downregulated. The most significantly up- or downregulated genes fell into two main categories, either associated with cell survival or with inflammation. This included genes such as ICOS (FDR p = 0.024) and S1PR1 (FDR p = 0.019) that were both up-regulated, indicating that a pro-inflammatory state is sustained in Long COVID PBMCs compared with COVID recovered PBMCs. Functional enrichment analysis identified that immune-related functions were expectedly predominant among the up- or down-regulated genes. The most frequently downregulated genes in significantly altered functional categories were two leukocyte immunoglobulin like receptors LILRB1 (FDR p = 0.005) and LILRB2 (FDR p = 0.027). PCA analysis demonstrated that LILRB1 and LILRB2 expression discriminated all of the Long COVID samples from COVID recovered samples.
Discussion: Downregulation of these inhibitory receptors similarly indicates a sustained pro-inflammatory state in Long COVID PBMCs. LILRB1 and LILRB2 should be validated as prospective biomarkers of Long COVID in larger cohorts, over time and against clinically overlapping conditions.
#mask up#public health#wear a mask#pandemic#wear a respirator#covid#covid 19#still coviding#coronavirus#sars cov 2#long covid#covid is not over
133 notes
·
View notes
Text
Inflamed from Within: How COVID-19 Ignites Heart Damage
Here is a version non-medical language terms.
Scientists recently uncovered something unsettling in a study of 54 heart tissue samples. The virus behind COVID-19 can creep into heart cells too, stirring up a storm of inflammation. These heart cells, called *cardiomyocytes*, are meant to keep our hearts beating strong, but this virus has found a way in, using the *TNF-NF-κB* pathway—a process that, when pushed too far, spells trouble.
Once inside, the virus seems to change the very way these heart cells function, flipping genetic switches that can lead to chaos. One gene, called *CXCL2*, kicks into high gear, summoning immune cells to the heart like soldiers to a battlefield. But sometimes, too many soldiers can cause more damage than they fix.
The protein at the center of this, *NF-κB*, is like the conductor of this chaotic orchestra, fueling inflammation that, if left unchecked, can weaken the heart. It’s as if the body, in trying to defend itself, starts tearing at its own foundation.
This study adds to a growing concern—COVID-19 is leaving its mark on the heart, and some of that damage may linger long after the virus has left. Scientists are sounding the alarm, and they’re urging us to take this more seriously than ever before. Posted by David It Up on X
39 notes
·
View notes
Text

Cardiovascular Variety
This study characterises the different cell types in different regions of the rat cardiovascular system using single-nucleus RNA sequencing (snRNA-seq) – 26 cell types are identified plus further subtypes. This image shows cell types in the aorta that were identified by snRNA-seq visually validated by immunofluorescence microscopy
Read the published research article here
Image from work by Alessandro Arduini, Stephen J. Fleming and Ling Xiao, and colleagues
Precision Cardiology Laboratory, The Broad Institute, Cambridge, MA, USA
Image originally published with a Creative Commons Attribution – NonCommercial – NoDerivs (CC BY-NC-ND 4.0)
Published in Cell Reports, January 2025
You can also follow BPoD on Instagram, Twitter and Facebook
10 notes
·
View notes
Note
Hey Meg! I was just catching up on your posts and wow, congrats on publishing the paper! I'm so excited for you and how far you'll go (it's so cool that you're working with alpha fold!!)! 🥳 How did you learn bioinformatics? Are there any resources you'd recommend for a complete beginner? (I'm trying to learn how to do proteomics analyses rn, but I can't seem to find many resources I can easily understand 🙈)
hii!! thank you so much for the wishes! the paper was a part of my final project with my team + guide so it was really a group effort <3 my teammates and I are all first authors, they were just kind enough to put my name first xD and yes! my current alphafold work is a lot of fun ^=^ I'm really enjoying my internship hehe
okay so I learnt bioinformatics as a part of my uni curriculum, and we mainly relied on these two textbooks
Essential Bioinformatics by Jin Xiong
Bioinformatics: Sequence and Genome Analysis
In addition to this, I also recently found this textbook titled Bioinformatics: An Introductory Textbook by Thomas Dandekar, Meik Kunz which focuses both on basic bioinformatics concepts and how to use the available tools. I especially like this book because I found it super easy to understand
EMBL also offers courses! even for synchronous online/in person courses that have ended, they tend to upload course materials. There's one for proteomics analyses as well- I've checked out the mass spectrometry videos under this course and they start with the very basics
In addition to these, here are some playlists on youtube that've helped:
Foundations of Systems and Computational Biology, MIT OpenCourseWare // classroom videos, I'd suggest to check specific videos for a topic of your choice rather than actually going through all of them
Computational Systems Biology, IITM // videos on the basics of systems biology, networks and modelling
Bioinformatics 101 // videos on how to use a lot of tools, however this primarily focuses on gene expression and RNA seq data
lastly, I don't know if this would be particularly helpful given what you're looking for, but this textbook on Computational Genomics with R is a blessing for Omics related Data Analysis. So if you ever need to use R in a biological context, this would be a great place to start
yeah! so these are the majority of the resources that I've used, I hope they help! Just a heads up that the links to the textbooks I mentioned in the beginning are direct download links and won't take you to any website.
If there's anything else I can help with, feel free to reach out ^=^ best of luck!
#if anyone has come across any helpful resources for bioinformatics feel free to add to this post...#so much of academia is hidden behind a paywall it's really a shame#I hope these will be of some help!#i don't use coursera/edx/etc so I didn't add any of those links on here#studyblr#resources#answered#bioinformatics#bio student#biology#regarding cpb
30 notes
·
View notes
Text
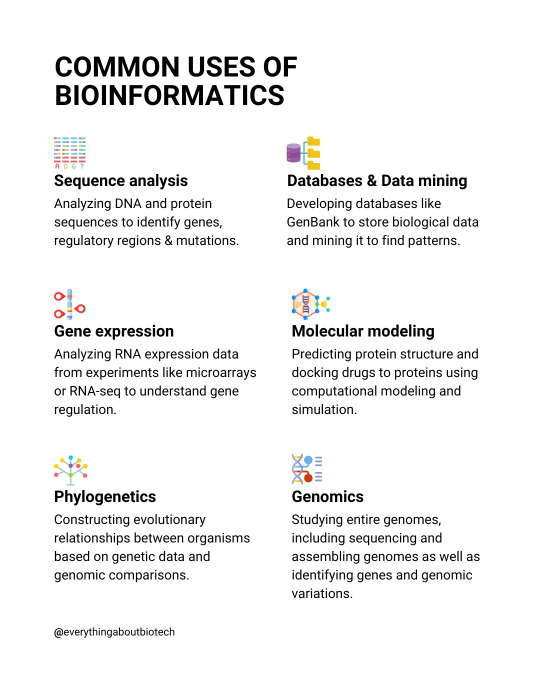
Common uses of bioinformatics
💡Sequence analysis Analyzing DNA and protein sequences to identify genes, regulatory regions & mutations.
💡Gene expression Analyzing RNA expression data from experiments like microarrays or RNA-seq to understand gene regulation.
💡Phylogenetics Constructing evolutionary relationships between organisms based on genetic data and genomic comparisons.
💡Molecular modeling Predicting protein structure and docking drugs to proteins using computational modeling and simulation.
💡Databases & Data mining Developing databases like GenBank to store biological data and mining it to find patterns.
💡Genomics Studying entire genomes, including sequencing and assembling genomes as well as identifying genes and genomic variations.
Follow @everythingaboutbiotech for useful posts.
#bioinformatics#genomics#proteomics#sequencing#PCR#biodata#bioIT#precisionmedicine#digitalhealth#biotech#DNA#healthtech#medtech#biostatistics#bioinformaticsjobs#BLAST#microarray#GenBank
54 notes
·
View notes
Text
Trinity is the most annoyingly touchy software package I swear to god. Take my god damn rna-seq files 🔫🔫🔫
#shout out to that one woman who commented on a google group in 2017 for saving me hours of work#bc literally why cant trinity understand sra files when literally everything else can#hisat2 didn't get mad at me trimmomatic didn't get mad at me
2 notes
·
View notes
Text
Antarctic moss proves cold weather isnt just for penguins
Antarctic moss proves cold weather isn’t just for penguins https://ift.tt/LciIb3E Mosses, which often go unnoticed in gardens and forests, have demonstrated an outstanding ability to adapt to extreme environments. Among them, Antarctic mosses are particularly interesting due to their resilience in one of the most challenging climates on Earth. Despite the freezing temperatures, prolonged drought, and intense ultraviolet radiation, these plants not only survive but also exhibit active growth during a brief summer season. Now, new research published in AoB PLANTS, shows that some Antarctic mosses adapt to these conditions by increasing the expression of genes related to lipid metabolism and the accumulation of unsaturated fatty acids. In this new study, the research team from Niigata University in Japan, focused on Bryum pseudotriquetrum, one of the more common mosses found in Antarctica. By employing transcriptome analyses, they explored how this species adapts to its challenging surroundings, particularly in comparison to controlled artificial conditions at 15°C. The research generated 88,205 contigs through de novo assembly, which represent the diverse array of genes expressed by this moss species. The analysis unveiled that under natural Antarctic field conditions 1,377 genes were upregulated, while 435 were downregulated when compared to those grown in more temperate, artificially controlled settings. But what does this mean for the moss’s survival? The upregulated genes included several related to lipid metabolism and the formation of oil bodies, two critical components that play a vital role in plants’ ability to cope with stress. Lipid metabolism is crucial for plants, especially in extreme environments where maintaining cellular integrity is essential. The study found that the expression levels of these lipid-related genes increased significantly in response to various artificial stress treatments, including low temperatures, salt exposure, and osmotic stress. This suggests that Bryum pseudotriquetrum has evolved mechanisms to change its lipid profiles in response to environmental challenges. Interestingly, the researchers observed that mosses grown in Antarctic conditions contained elevated levels of fatty acids, particularly α-linolenic acid, linolenic acid, and arachidonic acid and a higher proportion of unsaturated fatty acids. These fatty acids play a key role in maintaining membrane fluidity, and thus support cellular function in cold environments where membranes might otherwise become rigid. In the present study, RNA-seq analysis was carried out in the common moss Bryum pseudotriquetrum and genes related to lipid metabolism and oil body formation were found to be highly expressed in field samples. In plant cells, lipid accumulation and changes in fatty acid composition are important mechanisms for acquiring environmental stress tolerance. Thus, these genes may be involved in multiple stress tolerance in Bryum pseudotriquetrum growing in Antarctica. This study marks a significant advance in our understanding of how some plants respond to the extreme conditions of their native habitats. By providing the first gene expression profiles for mosses grown directly under Antarctic field conditions, it highlights the role of lipid metabolism and fatty acid composition in stress tolerance. The results also highlight the importance of in situ studies for understanding the mechanisms of plant resilience in stressed environments. READ THE ARTICLE Otani N., Kitamura H., Kudoh S., Imura S. and Nakano M. (2024) “Transcriptome analysis of the common moss Bryum pseudotriquetrum grown under Antarctic field condition” AoB PLANTS. Available at: https://doi.org/10.1093/aobpla/plae043 Cover image by HermannSchachner – Own work, CC0, Link The post Antarctic moss proves cold weather isn’t just for penguins appeared first on Botany One. via Botany One https://botany.one/ October 23, 2024 at 03:30PM
2 notes
·
View notes
Text
When I get bottom surgery, I genuinely want to ask them if I can keep some samples of testicular tissue, and use whatever lab resources are available to me at the time to do some extremely basic experiments.
At the very, VERY least, I would want any excess tissue homogenized and deep frozen for RNA-seq. If possible, it would also be cool to have some fixed tissue that would let me do IHC for testicular and ovarian enriched proteins. Even if full transdifferentiation of cells isn't possible based on HRT alone, in theory hormonal signaling should be driving expression of some of its downstream targets in primary sex organs, depending on how much it can reverse a lot of the cell fate "locks". Literally just a bulk RNA-seq that I could compare with sequences of cis male testicular tissue would be so fucking fascinating.
But who knows what I'll be doing when I actually get those surgeries and if they'd let me do something like that LOL
150 notes
·
View notes
Text
Navigating the Complexity of Alternative Splicing in Eukaryotic Gene Expression: A Molecular Odyssey
Embarking on the journey of molecular biology exposes students to the marvels and intricacies of life at the molecular level. One captivating aspect within this domain is the phenomenon of alternative splicing, where a single gene orchestrates a symphony of diverse protein isoforms. As students grapple with questions related to this molecular intricacy, the role of a reliable molecular biology Assignment Helper becomes indispensable. This blog delves into a challenging question, exploring the mechanisms and consequences of alternative splicing, shedding light on its pivotal role in molecular biology.
Question: Explain the mechanisms and consequences of alternative splicing in eukaryotic gene expression, highlighting its role in generating proteomic diversity and the potential impact on cellular function. Additionally, discuss any recent advancements or discoveries that have provided insight into the regulation and functional significance of alternative splicing.
Answer: Alternative splicing, a maestro in the grand composition of gene expression, intricately weaves the fabric of molecular diversity. Mechanistically, this phenomenon employs exon skipping, intron retention, and alternative 5' or 3' splice sites to sculpt multiple mRNA isoforms from a single gene.
The repercussions of alternative splicing resonate deeply within the proteomic landscape. Proteins, diverse in function, emerge as a consequence, adding layers of complexity to cellular processes. Tissue-specific expression, another outcome, paints a vivid picture of the nuanced orchestration of cellular differentiation.
Regulating this intricate dance of alternative splicing involves an ensemble cast of splicing factors, enhancers, silencers, and epigenetic modifications. In the ever-evolving landscape, recent breakthroughs in high-throughput sequencing techniques, notably RNA-seq, offer a panoramic view of splicing patterns across diverse tissues and conditions. CRISPR/Cas9 technology, a molecular tool of precision, enables the manipulation of splicing factor expression, unraveling their roles in the intricate regulation of alternative splicing.
In the dynamic realm of molecular biology, alternative splicing emerges as a linchpin. Specific splicing events, linked to various diseases, beckon researchers towards therapeutic interventions. The complexities embedded in this molecular tapestry underscore the perpetual need for exploration and comprehension.
Conclusion: The odyssey through alternative splicing unveils its prominence as a cornerstone in the narrative of molecular biology. From sculpting proteomic diversity to influencing cellular functions, alternative splicing encapsulates the essence of molecular intricacies. For students navigating this terrain, the exploration of questions like these not only deepens understanding but also propels us into a realm of limitless possibilities.
#molecular biology assignment help#biology assignment help#university#college#assignment help#pay to do assignment
9 notes
·
View notes
Text
COVID-19 patients showed an impaired ability to kill intracellular bacteria, which could increase the risk of bacterial superinfections.
The presence of intracellular bacteria affected the immune cell functions and interactions with the virus.
5 notes
·
View notes
Text

the casual violence of covid is such that this is supposed to be reasonable to me, someone with severe me/cfs trapped in my house with my dad who doesn't wear a fucking mask and my mom and partner and I all taking precautions and always masking when we go out.
my blood boils constantly. the vast majority of what makes up my life has already been taken from me. this is so violent and I'm just supposed to take it. and of course I don't want my mom or partner to get this either, it's so bad! (my dad can go fuck himself at this point)
this is the kind of shit covid can do to you:
covid damages your body with each infection. it wears down your immune system and damages t cells in a way similar to HIV. I am this sick because of postviral illness after a mono infection I got about 10 years ago. I am lucky if I get a few hours a day I can do anything at all. I am scared if I get long covid that I may die or have such poor quality of life that I won't want to live.
if you are masking and being cautious about covid while being put into fucked up situations you're not alone and it's not okay that this is happening to us. the grief and rage is real. we are not disposable. we are people. we matter.
#personal#chronic illness#disability#ableism#me/cfs#chronic fatigue syndrome#chronically ill#disabled#covid
3 notes
·
View notes