#m&gedit
Text
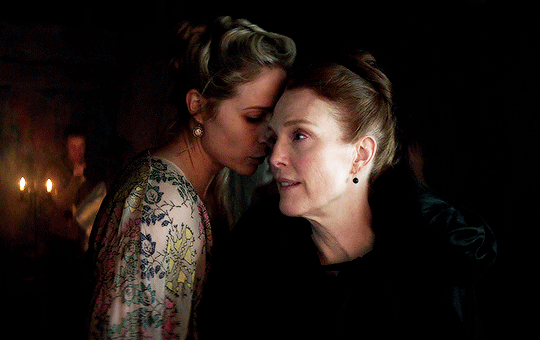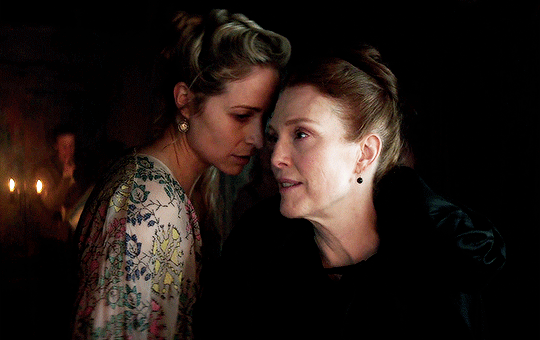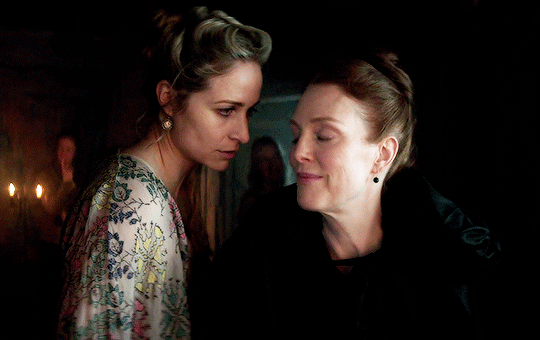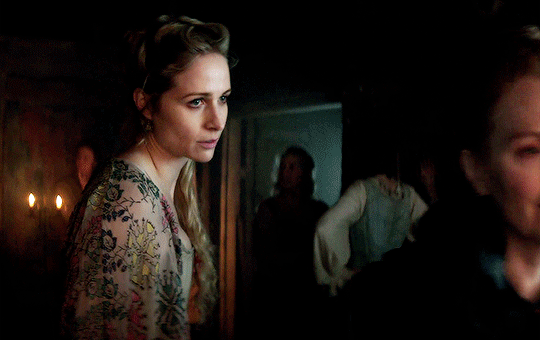
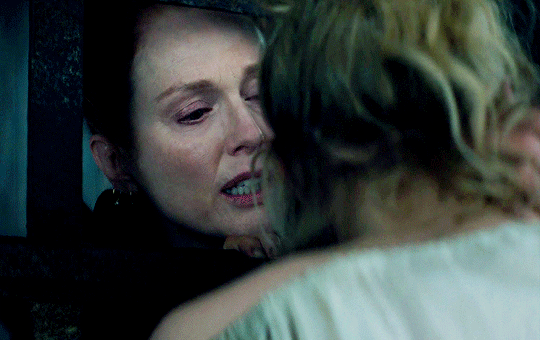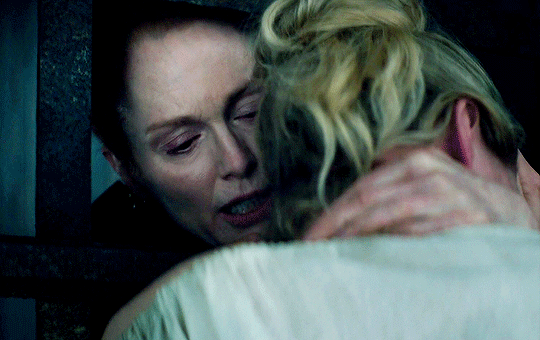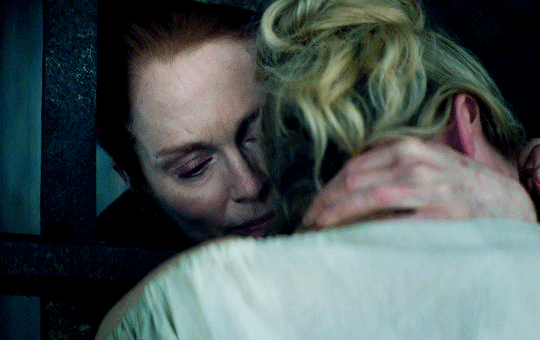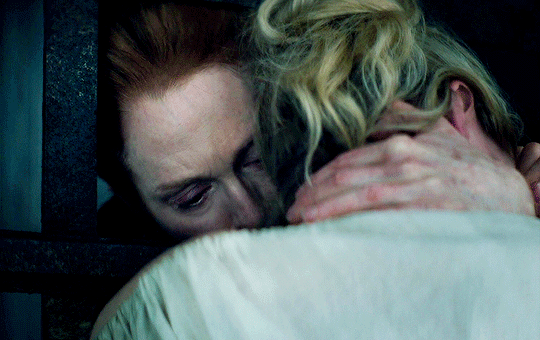

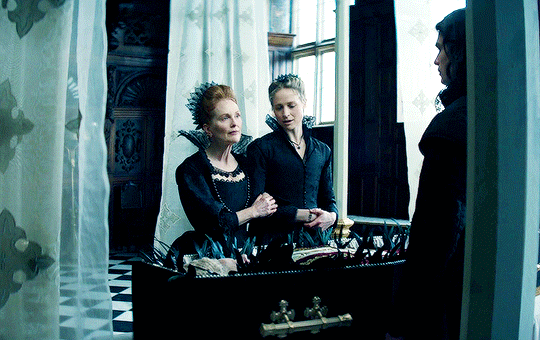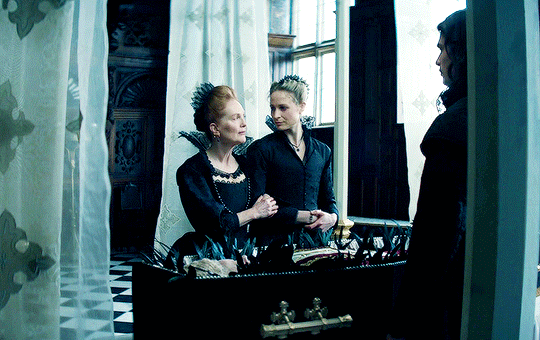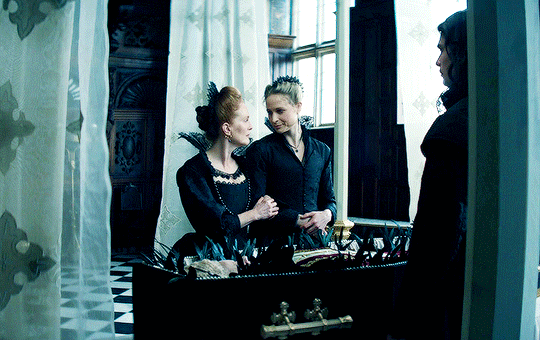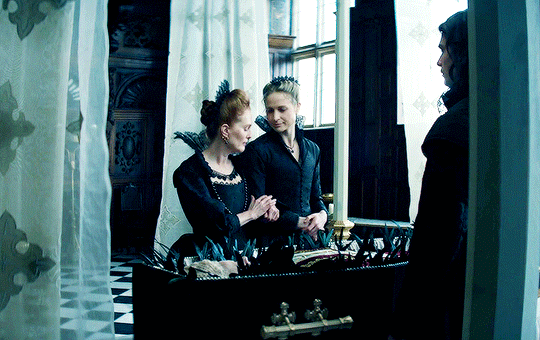



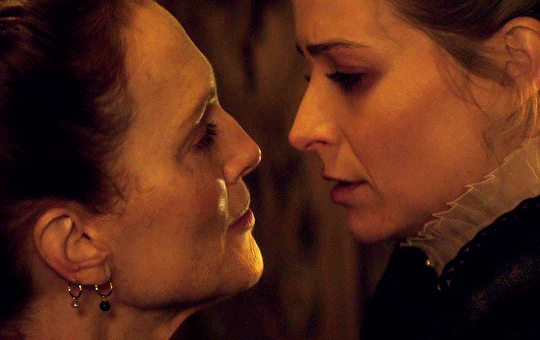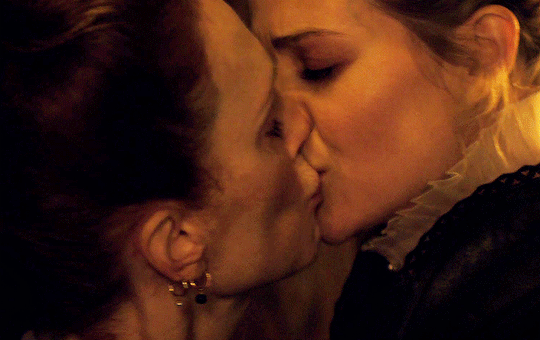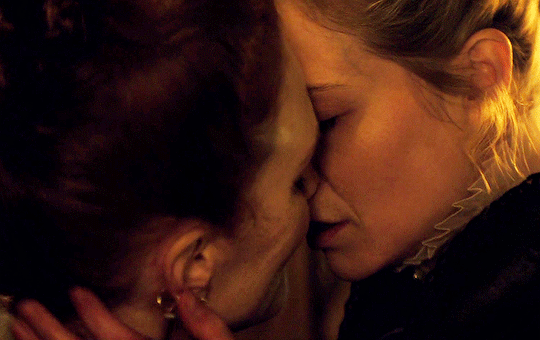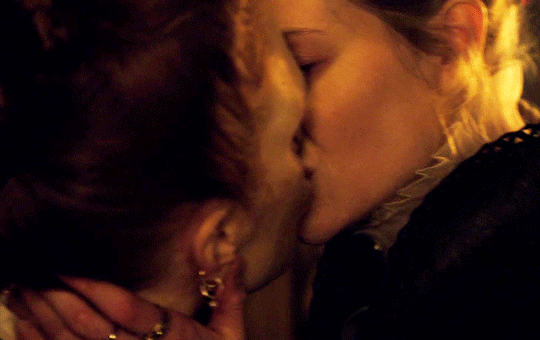

I vow to you, as I battle, you will rise.
MARY VILLIERS and SANDIE BROOKS
Mary & George
#mary x sandie#mary & george#mary villiers#sandie brooks#julianne moore#niamh algar#wlwgif#wlwedit#m&gedit#m&g spoilers#my edits
1K notes
·
View notes
Text


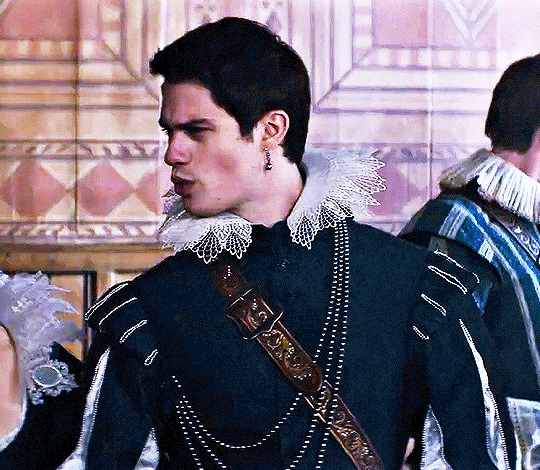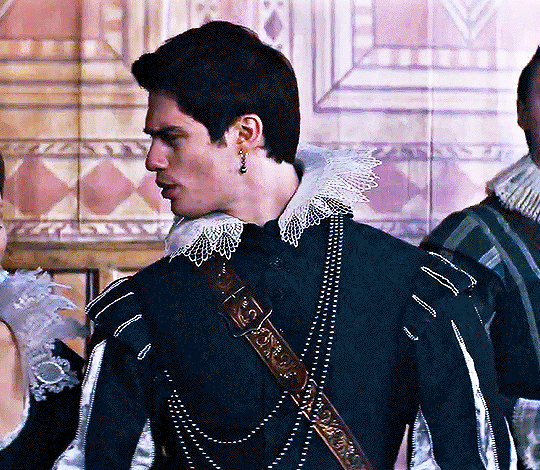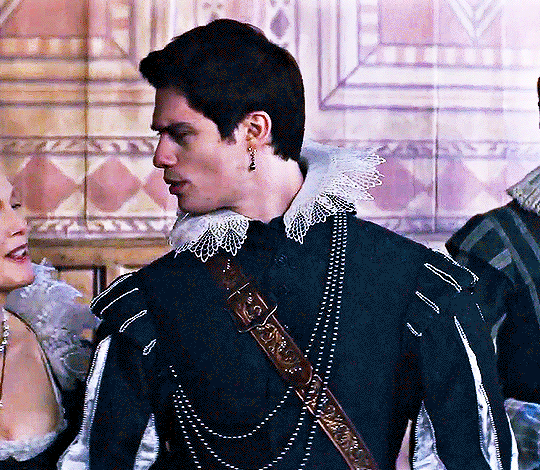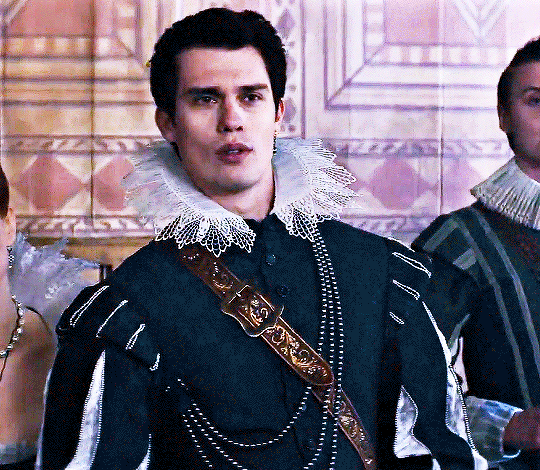
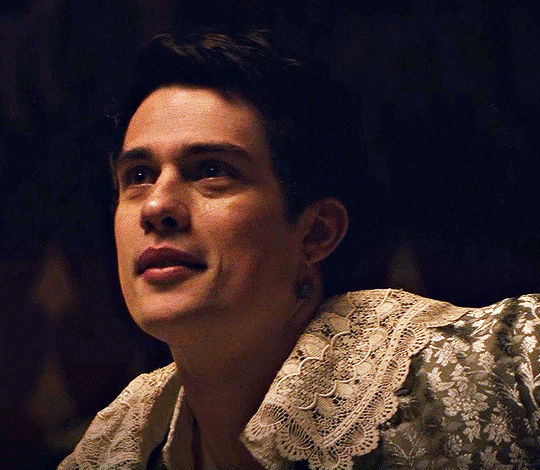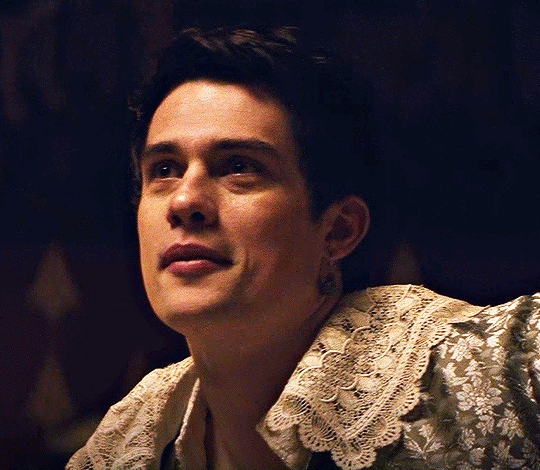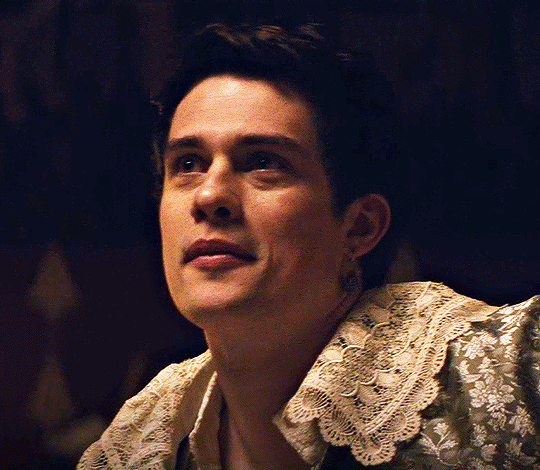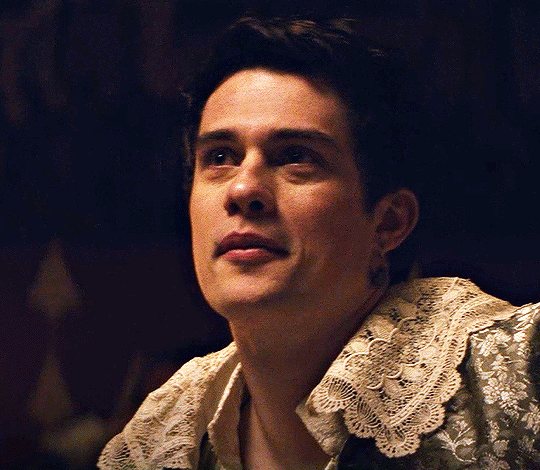

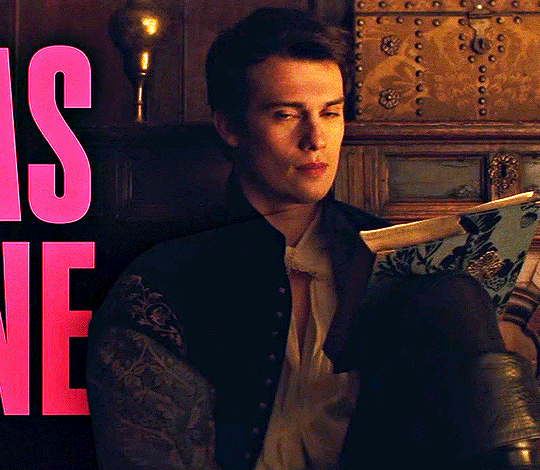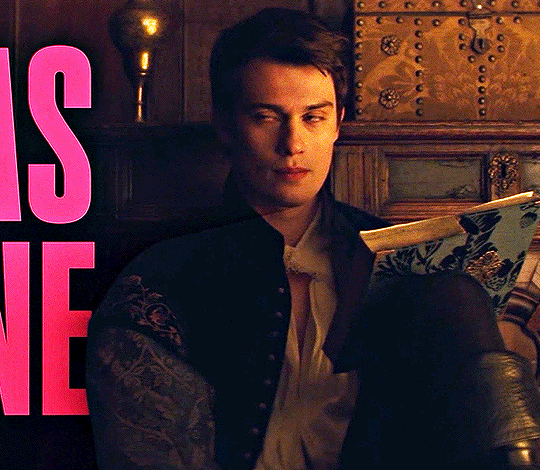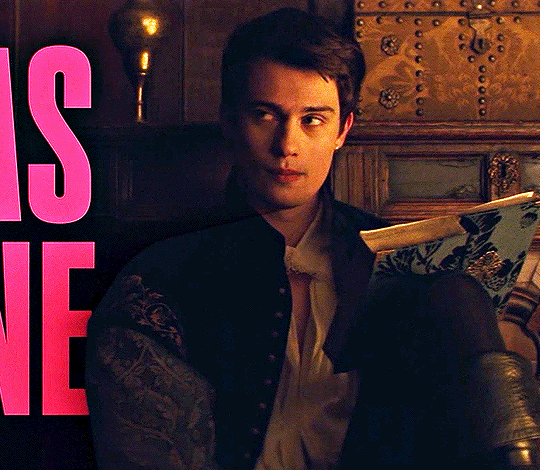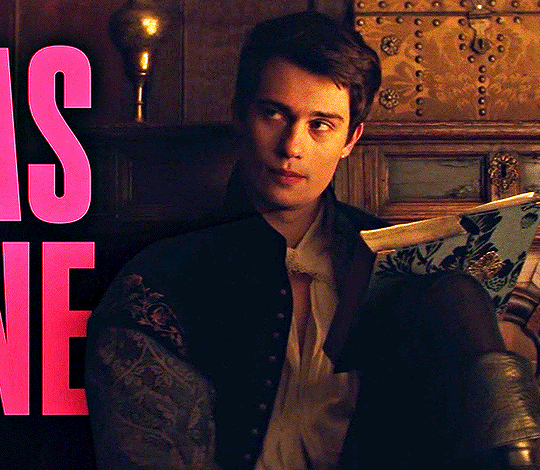


NICHOLAS GALITZINE as GEORGE VILLIERS
Mary & George // Official Trailer
#mary & george#mary and george#m&gedit#nicholas galitzine#perioddramaedit#maryandgeorgeedit#userninz#chrissiewatts#usersteen#usernuria#usermegsb#mine*#HELLOOOOOO GEORGE VILLIERS NATION ARE WE UP#nick is going to eat up this role sooooo much#im so excited for this#the costumes...... woah.#idk who else is watching this but lmk if youre up for tagging :D (or not)
959 notes
·
View notes
Text



put your head on my chest.
#maryandgeorgeedit#mary and george#nicholas galitzine#tony curran#m&gedit#george villiers#king james
385 notes
·
View notes
Text






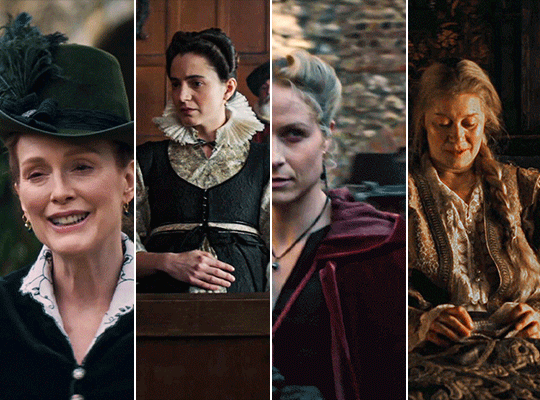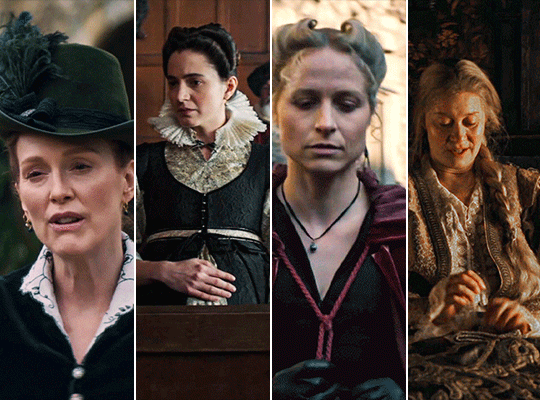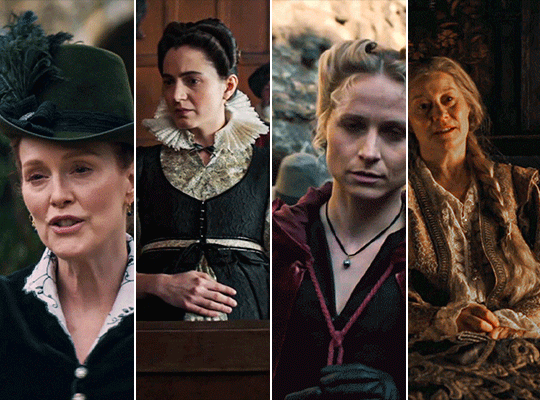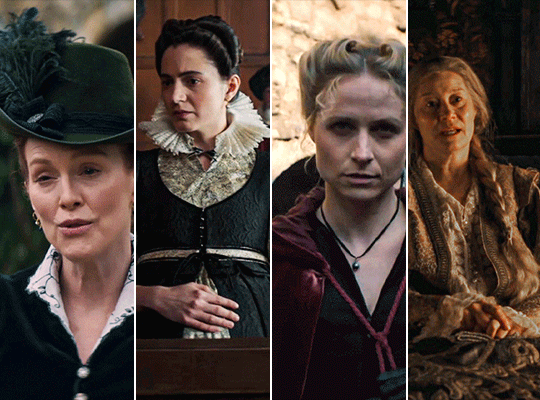
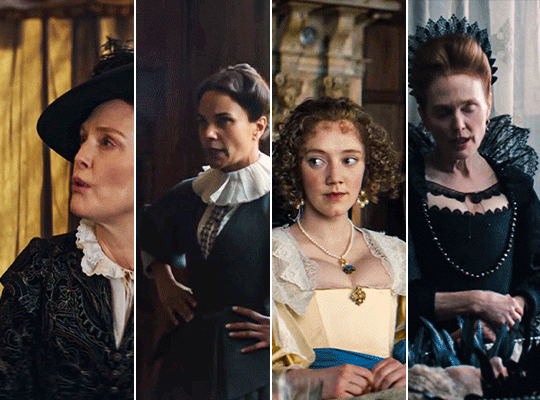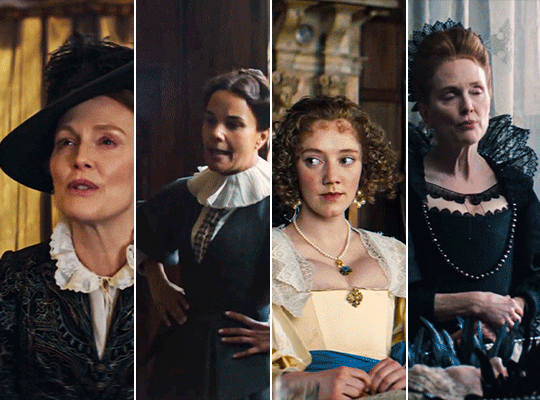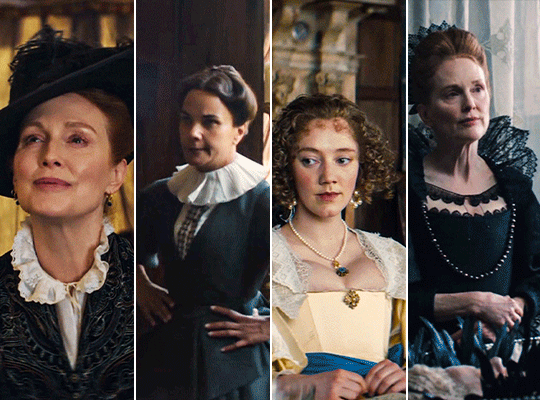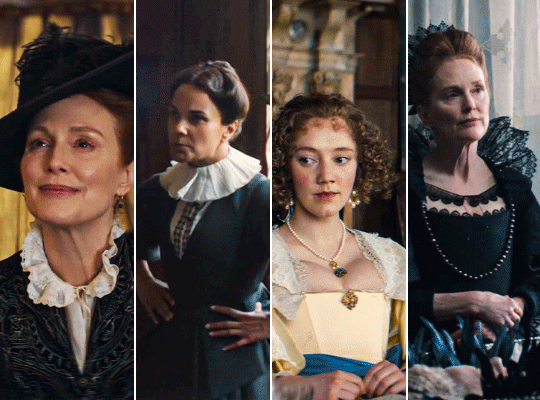
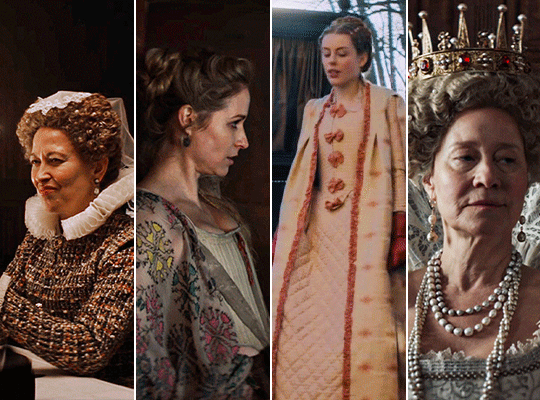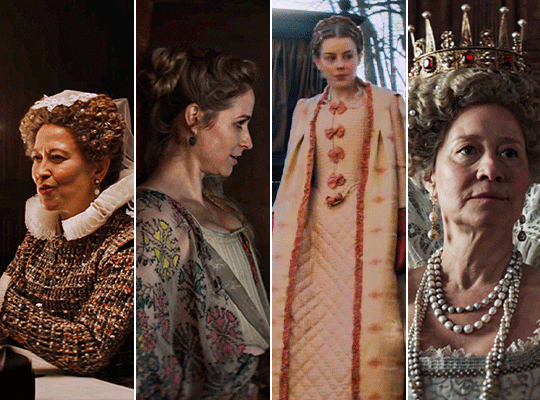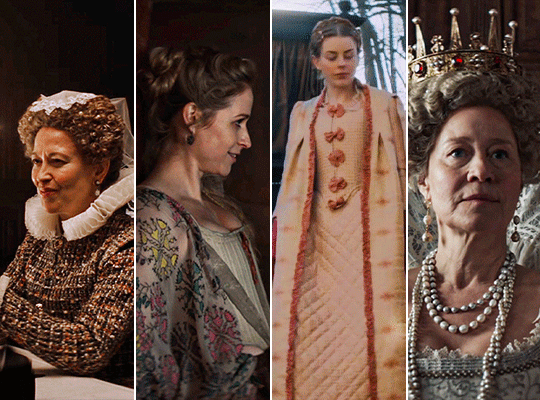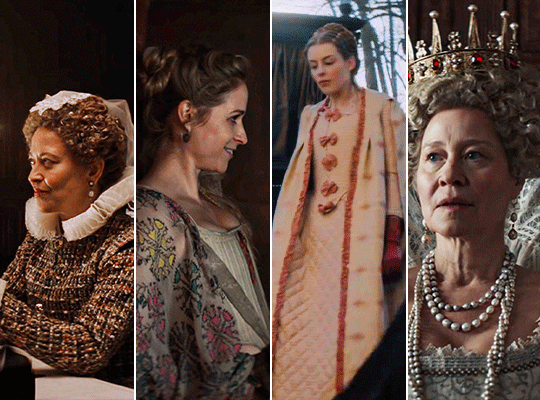

MARY & GEORGE (2024) → costume design annie ruth symons
#mary and george#mary & george#mary villiers#julianne moore#niamh algar#maryandgeorgeedit#m&gedit#userstream#perioddramaedit#perioddramasource#dailyfilmtvgifs#swearphil#usersteen#chrissiewatts#iuserzoe#userbeckett#*#my edit#oh my god this took my entire day#i'm gonna go play spider-man
264 notes
·
View notes
Text






NICHOLAS GALITIZINE in MARY & GEORGE
ep 1x01: THE SECOND SON
#lowq mb...#mary & george#nicholas galitzine#george villiers#tvedit#ngalitzineedit#m&gedit#m*#m&g*#i stand w my bi wife!
150 notes
·
View notes
Photo

Yeni yazımıza göz atın https://www.teknoloskop.net/linuxta-c-programlama-nasil-yapilir/
Linux'ta C Programlama Nasıl Yapılır?

Dünya’da en aktif kullanılan programlama dillerinden olan ve birçok işletim sistemine tabanlık yapan C, Linux tabanlı sistemlerde nasıl kodlanır, derlenir ve çalıştırılır?
1972 yılında Dennis M. Ritchie tarafından kullanıma yayımlanan ve Dünya’da programcılık dillerinden bir kısmını etkilen C Programla Dili bilgisayar mühendisleri ve programcıları için çok önemli bir dildir. Dünya üzerinde aktif olan kullanılan birçok sistem kendine taban olarak C Programlama Dilini seçmiştir. Ayrıca mühendisler ve programcılar tarafından aktif olarak kullanılan C#, PHP, JavaScript, Java ve ASP de C Programlama Dilinden direkt ya da dolaylı olarak etkilenmiş, kendilerine ait bir dil sunmuşlardır. UNIX işletim sisteminin üretiminde de C Programlama Dili kullanılmıştır. Günümüzde en aktif olarak kullanan ve birçok kullanıcıya erişen Apple‘ın üretimi Mac OS X de C Programlama Dilini taban alır.
Linux’ta C Programlamaya giriş
Linux çekirdekli bilgisayarlarda C Programlama dilini yazdıktan sonra işlemler yapabilmek için açık kaynaklı olan C tabanını sistemimize kurmamız gerekiyor. Bu kuracağımız paket sayesinde sistemimiz C Programla kodlarını compile (inşa) edebilecek ve kodlama ile sağladığımız girdileri çalıştırabilecektir. Bu paket C Programlama Dilinin kullanımı amacıyla bizzat Özgür Yazılım Vakfı (GNU) tarafından üretiliyor. Eğer bu pakete işletim siteminiz sahip değilse işletim sisteminiz C Programlama Dili ile yazılan kodları tanımlayamaz ve bir execution (çalışma-yürütüm) elde edemezsiniz.
İşletim sisteminizi C Progamlama Diline hazırlamak için ilk önce aşağıdaki komutu uçbirimde çalıştırmanız gerekiyor.
Debian tabanlı sistemler için girilecek kod;
sudo apt-get install gcc
Arch tabanlı sistemleri için girilecek kod;
pacman -S gcc
İkinci Adım
Yukarıdaki komutların girdisi ile kurulan paketlerin sonunda başarılı bir sonuç aldıysanız artık işletim sisteminiz C Programla Dili ile yazılan girdileri inşa edip yürütebilecek. Artık C Kodlarını yazmaya başlayabilirsiniz. Kodlarınızı isterseniz gedit gibi bir metin düzenleyicide ya da kodlama, inşa ve yürütme işlemlerini yapabileceğiniz bir yazılım kurabilirsiniz. Bu noktada size yardımcı bir yazılım öneriyoruz çünkü hata yapıldığı zamanlarda, yardımcı program hatalarınız nerede olacağını bildirecek ve hatanızı aramakla zaman kaybetmeyeceksiniz.
Size kurmanızı önereceğimiz yazılım ise Geany. Geany sadece C Programla Dili değil birçok programlama dilini destekleyen program. Eğer başka bir programlama diliyle de ilgileniyorsanız Geany ile iki yardımcı yazılım yerine tek bir yardımcı yazılım kullanabilirsiniz.
Geany Kurulumu
Dilerseniz işletim sisteminizin Uygulama Yöneticisinden Geany’i kurabilir ya da komut satırından kısaca bir komut girerek Geany’i kurabilirsiniz. Geany’i komut satırından komut girerek kurmak isterseniz aşağıdaki komutları işletim sisteminizin komut satırına giriniz.
Debian tabanlı işletim sistemlerinde Geany kurmak için komut
apt-get install geany
Arch tabanlı işletim sistemlerinde Geany kurmak için komut
pacman -S geany
Komutların girişinden sonra başarılı bir kurulum gerçekleştirirseniz artık bilgisayarınız ve dolaylı olarak işletim sisteminiz C Programlama Dilini inşa edebilir ve kodların girdilerini yürüterek size bir çıktı sağlayabilir.
Örnek bir C Programlama Kodu
Kurduğumuz yardımcı yazılım Geany’i denemek için basit bir kod yazalım. Kod ile ekrana “Teknoloskop ile C Programlama yapacağım.” yazsın. Bunun için aşağıdaki kodu Geany’de yeni bir C dosyası açarak giriniz. Bu kod yazma işlemini de adım adım yani satır satır uygulayalım.
#include
int main (void)
printf(“Teknoloskop ile C Programlama yapacağım.”);
Artık bir bilgisayar programcısısınız. Tebrikler.
0 notes
Photo

VIMKiller
VI(M) is hard, and sometimes we need to take drastic measures. We understand your needs. Maybe you're new on the job, and you forgot to set your default text editor to nano, emacs, gedit, whatever. VIM pops up and now you have to make a choice...
Read
0 notes
Text
Computers in the German Democratic Republic
New Post has been published on https://www.aneddoticamagazine.com/ddr-computer/
Computers in the German Democratic Republic

DDR Deutsche Demokratische Republik
PIKO dat Computer 1969
youtube
PIKO dat Computer Program v1
youtube
PIKO dat Computer build in DDR in 1969. Demo of how to build a program by using cable bridges.
LC 80 (1982)
Lerncomputer
VEB Kombinat Robotron was the biggest East German electronics manufacturer. It was based in Dresden and employed 68,000 people (1989). It produced personal computers, SM EVM minicomputers, the ESER mainframe computers, several computer peripherals as well as home computers, radios and television sets.
Robotron managed several different divisions:
VEB Robotron-Elektronik Dresden (headquarters) — typewriters, personal computers, minicomputers, mainframes
VEB Robotron-Meßelektronik Dresden — measurement and testing devices, home computers
VEB Robotron-Projekt Dresden — software department
VEB Robotron-Buchungsmaschinenwerk Karl-Marx-Stadt — personal computers, floppy disk drives
VEB Robotron-Elektronik Hoyerswerda — monitors, power supply units
VEB Robotron-Elektronik Radeberg — radio receivers, portable television receivers
VEB Robotron Vertrieb Dresden, Berlin and Erfurt — sales departments
VEB Robotron-Elektronik Zella-Mehlis — computer terminals, hard disk drives
VEB Robotron-Büromaschinenwerk Sömmerda — personal computers, printers, electronic calculators (Soemtron 220, 222, 224), invoicing machines (EFA 380), punched card indexers and sorters (Soemtron 432).
VEB Robotron Elektronik Riesa — printed circuit boards
On 30 June 1990 the Kombinat Robotron was liquidated and the divisions were converted into corporations. In the 1990s these companies were sold, e.g., to Siemens Nixdorf, or also liquidated.
Robotron A 5120 office computer, 1982
Licensed under CC BY-SA 2.5 via Wikimedia Commons.
KC 87 (1984)
KC 85/4 (1998)
KC compact (1989)
P8000
CPU 8 bit: UA880 (2 EPROM, 8*64K bit DRAM U264 u.a.)
CPU 16 bit: UB8001 (4 EPROM u.a.)
Taktfrequenz 4MHz
Speichererweiterung: 256 Kbyte RAM
Peripherie: 2*5,25 Zoll Floppy Disk integriert, Terminal Beistellgerät mit 2*5,25 oder 8 Zoll
Interface: 8* V24 Schnittstellen
Betriebssystem für 16 Bit: WEGA, für 8 Bit: OS/M (CP/M 2.2, SCPX), IS/M (ISIS II), UDOS
Hersteller: Elektro-Apparate-Werke (=EAW) Berlin-Treptow
Robotron Computermuseum http://www.efb-1.de/
http://www.mobiltom.de/z1013.html
http://www.robotrontechnik.de/
Robotron A7100 with GEdit /M16 2D CAD Program CP/M-86 2.2
DDR Computer robotron A7100 16 Bit 8086-er laed das 2D CAD Programm “GEDIT /M16”.
Gleiches Spiel wie mit Grafik/M16:
* Start mit SCP1700 V.2.2 Bootdiskette: (CP/M-86 2.2 Derivat)
* Laden der KGS Firmware GRAF6.FRM in den RAM der KGS- Karte (grafisches SUB System).
Grafikkarte und KGS-Karte bilden eine Einheit und sind ueber eine Back Plane im Rechner miteinander verbunden.
Prozessor der Karten ist eine 8 Bit U880 CPU ein Z80 Clone von Zilog,
ueber den alle grafischen Befehle abgearbeitet werden.
* Laden der SCP-GX Schnittstellensteuerung (GRAPHICS.CMD), zum Umschalten in den grafischen Modus mit 640×400 Bildpunkten.
* Laden der Programmdatei: GEDIT.CMD
P8000 – Ein DDR Computer von 1988 in Aktion
youtube
P 8000 ist ein Rechner, der Ende der 80-er Jahre im EAW Elektronik (DDR) gebaut wurde. Es gab Varianten mit 8-bit und welche mit zusätzlichem 16-bit Teil.
Hier zu sehen ist die 8-bit Variante mit 64 kB RAM und zwei 5 1/4 Zoll Diskettenlaufwerken. Die Ein- und Ausgabe erfolgt über Terminals.
Der Rechner wurde hauptsächlich für wissenschaftlich-technische Zwecke und zur Programmierung genutzt.
Im Video läd er das Betriebssystem OS/M 3.0 (ein CP/M Clone) und danach die Statistik Software ABSTAT. Über den 9-Nadel A3 Drucker Robotron K 6314 erfolgt der Ausdruck der Daten.
P8000 Compact
DDR-Homecomputer Robotron KC87 Demoprogramm (80-er Jahre)
youtube
Basic Demoprogramm auf einem Robotron KC87, Stimme kommt von Programmkassette, Langfassung (80-er)
DDR Computer Robotron PC 1715 aus den 80-ern mit den Spielen Pilots und Leiter
youtube
Viele Bürocomputer der 80-er Jahre waren noch nicht grafikfähig. Um dennoch Spiele für diese Maschinen programmieren zu können musste man findig sein und aus den Standard-Textzeichen eine “Quasigrafik” (oder auch Businessgrafik genannt) zusammensetzten.
Hier zwei Bespiele: die Programme Pilots und Leiter. Insbesondere letzteres Spiel war auf den DDR Bürocomputern der 80-er Jahre häufig zu finden.
1988 wurde dieser CAD-Arbeitsplatz in der DDR entwickelt, der aus einem A7150 (CM1910), einem Farbmonitor, dem Grafiktablett K6405 und dem Plotter K6418 (CM6415) bestand.
Die Robotron CAD Software war GEDIT, es wurde aber auch mitunter ein angepasstes AutoCad benutzt.
Der Computer war bereits ein leistungsfähiger 16-bit Rechner mit 8086 Prozessor und Festplatte. Der CAD Arbeitsplatz lief unter SCP1700 (CP/M86) oder DCP (MS-DOS).
Der Arbeitsplatz ist ein Exponat des Robotron-Computermuseums www.efb-1.de
ddr geldautomat
youtube
Werbung für Geldautomaten in der DDR. Es handelt sich hier um ein Basic-Programm, welches auf einem KC85/4 abläuft. Diese Automaten wurden Ende der 1980er Jahren, so ab etwa 1988 in einigen Städten aufgestellt. näheres zu diesen Geräten unter www.robotrontechnik.de
Robotron – Computer made in GDR
youtube
Komplizierte Rechnungen finden heutzutage in Hosentaschen oder in Armbanduhren statt und die kleinen technischen Wunderwerke, die sie ausführen, sind aus dem täglichen Leben nicht mehr wegzudenken. Der Beginn der digitalen Revolution ist in der DDR mit dem Namen ROBOTRON fest verbunden. Der Chemnitzer Rolf Kutschbach gilt als der Vater der Rechentechnik in der DDR. In “GMD – Das Magazin” erzählt er von der abenteuerlichen Entwicklung des “R300” und verrät, warum der Rechner trotz internationaler Anfragen nicht in den Export kam.
Gekürztes Demonstrationsprogramm auf einem Robotron A5105 (BIC – Bildungscomputer) , geschrieben in BASIC. (1989)
DDR-Bildungscomputer Robotron A5105 – BIC Demo-Programm (80-er Jahre)
youtube
Robotron Typenraddrucker SD1152 in Aktion (Daisywheelprinter)
youtube
Robotron CAD-Arbeitsplatz mit CM1910 (A7150), Plotter CM6415 (K6418) und Grafiktablett K6405
youtube
1988 wurde dieser CAD-Arbeitsplatz in der DDR entwickelt, der aus einem A7150 (CM1910), einem Farbmonitor, dem Grafiktablett K6405 und dem Plotter K6418 (CM6415) bestand.
Die Robotron CAD Software war GEDIT, es wurde aber auch mitunter ein angepasstes AutoCad benutzt.
Der Computer war bereits ein leistungsfähiger 16-bit Rechner mit 8086 Prozessor und Festplatte. Der CAD Arbeitsplatz lief unter SCP1700 (CP/M86) oder DCP (MS-DOS).
Der Arbeitsplatz ist ein Exponat des Robotron-Computermuseums www.efb-1.de
Mansfeld Prozess Controller ( MPC ), ein 8-bit DDR-Computer in Aktion
youtube
DDR-Homecomputer Robotron KC87 Demoprogramm (80-er Jahre)
youtube
Nach 22 Jahren und 400 Sendungen WDR Computerclub wurde das beliebte Computermagazin mit Wolfgang Back und Wolfgang Rudolph im Jahr 2003 abgesetzt. Aber als ComputerClub 2 lebt das Magazin auf NRW.TV und unter cczwei.de weiter.
DDR Computer Robotron PC 1715 aus den 80-ern mit den Spielen Pilots und Leiter
youtube
Viele Bürocomputer der 80-er Jahre waren noch nicht grafikfähig. Um dennoch Spiele für diese Maschinen programmieren zu können musste man findig sein und aus den Standard-Textzeichen eine “Quasigrafik” (oder auch Businessgrafik genannt) zusammensetzten.
Hier zwei Bespiele: die Programme Pilots und Leiter. Insbesondere letzteres Spiel war auf den DDR Bürocomputern der 80-er Jahre häufig zu finden.
Robotron now
youtube
401
0 notes
Text
Cómo instalar AdoptOpenJDK en las principales distribuciones
En el artículo donde explicamos la verdadera situación de Java (o mejor dicho el JDK) desde que se volvió de pago mencionamos una iniciativa llamada AdoptOpenJDK, la cual tiene el propósito de facilitar la difusión e instalación de OpenJDK sobre todo entre los usuarios de Windows y Mac, ya que aparentemente la mayoría de estos no conocían este último proyecto, a pesar de ser la base tecnológica de Java tal y como lo conocemos hoy en día.
Aunque AdoptOpenJDK iba dirigido sobre todo a los usuarios de Windows y Mac, la mayor difusión que ha tenido en los últimos tiempos derivó en la demanda por parte de los usuarios de GNU/Linux de un mejor soporte, automatizando el mantenimiento mediante repositorios oficiales en lugar de tener que actualizar manualmente la instalación manual (valga la redundancia) que se tenía que hacer a partir del fichero tar.gz.
Después de muchas peticiones, la iniciativa AdoptOpenJDK ha puesto hace poco a disposición repositorios oficiales que suministran paquetes Deb y RPM, soportando las principales distribuciones que utilizan dichos formatos de paquetes: Ubuntu (y toda derivada directa como Linux Mint o KDE neon), Debian, CentOS, RHEL, Fedora, openSUSE Leap y SLES. Tomando como referencia los comandos publicados por George Adams, vamos a mostrar la instalación en Ubuntu, Debian, Fedora 30 y openSUSE Leap 15.
Instalar AdoptOpenJDK en Ubuntu y Debian
Para configurar el repositorio de AdoptOpenJDK en Ubuntu y Debian solo hay que seguir el procedimiento normal para estos casos (en caso de no tener configurado “sudo” en Debian, hay que iniciar la sesión como root y luego realizar los comandos sin poner “sudo” delante).
Primero se obtiene la clave GPG del repositorio:
wget -qO - https://adoptopenjdk.jfrog.io/adoptopenjdk/api/gpg/key/public | sudo apt-key add -
Después de añade el repositorio:
sudo add-apt-repository --yes https://adoptopenjdk.jfrog.io/adoptopenjdk/deb/
Se refresca la paquetería:
sudo apt update
Instalando la versión 8 del JRE:
sudo apt install adoptopenjdk-8-hotspot-jre
Instalando la versión 11 del JRE:
sudo apt install adoptopenjdk-11-hotspot-jre
Instalando la versión 12 del JRE:
sudo apt install adoptopenjdk-12-hotspot-jre
Esquema de instalación del JRE (sustituir ‘<version>’ por la versión que se quiere instalar):
sudo apt install adoptopenjdk-<version>-hotspot-jre
Instalando la versión 8 del JDK (tendría que instalar el JRE como dependencia):
sudo apt install adoptopenjdk-8-hotspot
Instalando la versión 11 del JDK:
sudo apt install adoptopenjdk-11-hotspot
Instalando la versión 12 del JDK:
sudo apt install adoptopenjdk-12-hotspot
Esquema de instalación del JDK (sustituir ‘<version>’ por la versión que se quiere instalar):
sudo apt install adoptopenjdk-<version>-hotspot
Instalar AdoptOpenJDK en Fedora 30
En Fedora 30 la instalación tiene menos pasos que en Ubuntu/Debian. Los que quieran instalar AdoptOpenJDK en otro sistemas construidos con tecnologías de Red Hat pueden seguir el siguiente enlace y adaptar el comando de Fedora 30 a otro sistema (siguiente el ejemplo, escribir ‘centos’ o ‘rhel’ en lugar de ‘fedora’ y estableciendo un número de versión en caso de no funcionar la variable “$releasever”). También es importante tener en cuenta cambiar el comando DNF por Yum en caso de usarse RHEL.
Abrir el siguiente fichero en Fedora 30 Workstation con permisos de administrador (se puede cambiar Gedit por otro editor de texto gráfico como Geany o bien uno de consola como Nano o Vim):
sudo gedit /etc/yum.repos.d/adoptopenjdk.repo
Introducir el siguiente texto en el fichero, guardar y cerrar:
[adoptopenjdk] name=AdoptOpenJDK $releasever - $basearch baseurl=https://adoptopenjdk.jfrog.io/adoptopenjdk/rpm/fedora/$releasever/$basearch enabled=1 gpgcheck=1 gpgkey=https://adoptopenjdk.jfrog.io/adoptopenjdk/api/gpg/key/public
Instalar la versión 8 del JRE:
sudo dnf install adoptopenjdk-8-hotspot-jre
Instalar la versión 11 del JRE:
sudo dnf install adoptopenjdk-11-hotspot-jre
Instalar la versión 12 del JRE:
sudo dnf install adoptopenjdk-12-hotspot-jre
Instalando otra versión del JRE (cambiar ‘<version>’ por la versión que corresponda):
sudo dnf install adoptopenjdk-<version>-hotspot-jre
Instalar la versión 8 del JDK (tendría que instalar el JRE como dependencia):
sudo dnf install adoptopenjdk-8-hotspot
Instalar la versión 11 del JDK:
sudo dnf install adoptopenjdk-11-hotspot
Instalar la versión 12 del JDK:
sudo dnf install adoptopenjdk-12-hotspot
Instalando otra versión del JDK:
sudo dnf install adoptopenjdk-<version>-hotspot
En la primera instalación se pedirá al usuario aceptar la clave del repositorio, la cual obviamente se tiene que aceptar.
Las instrucciones publicadas por George Adams son para un contenedor Docker, así que he tenido que preguntar en el repositorio de GitHub de AdoptOpenJDK para obtener una configuración válida para un sistema Fedora “real” (escritorio o servidor).
Instalar AdoptOpenJDK en openSUSE Leap 15
La iniciativa AdoptOpenJDK también ofrece repositorios para openSUSE Leap 15.0 y SLES (SuSE Linux Enterprise Server). Aquí mostraremos los pasos para instalarlo en la distribución comunitaria, aunque solo habría que cambiar la referencia al sistema operativo (poniendo ‘sles’ en lugar de ‘opensuse’ y cambiando el ‘15.0’ por la versión que corresponda) en caso de usarse el sistema corporativo. Al igual que con las distribuciones de tecnologías Red Hat, en este enlace se pueden comprobar todas las distribuciones y versiones soportadas.
Configuración del repositorio en openSUSE Leap 15.0:
sudo zypper ar -f http://adoptopenjdk.jfrog.io/adoptopenjdk/rpm/opensuse/15.0/$(uname -m) adoptopenjdk
Instalar la versión 8 del JRE:
sudo zypper install adoptopenjdk-8-hotspot-jre
Instalar la versión 11 del JRE:
sudo zypper install adoptopenjdk-11-hotspot-jre
Instalar la versión 12 del JRE:
sudo zypper install adoptopenjdk-12-hotspot-jre
Instalando otra versión del JRE:
sudo zypper install adoptopenjdk-<version>-hotspot-jre
Instalar la versión 8 del JDK (tendría que instalar el JRE como dependencia):
sudo zypper install adoptopenjdk-8-hotspot
Instalar la versión 11 del JDK:
sudo zypper install adoptopenjdk-11-hotspot
Instalar la versión 12 del JDK:
sudo zypper install adoptopenjdk-12-hotspot
Instalando otra versión del JDK:
sudo zypper install adoptopenjdk-<version>-hotspot
¿Es necesario instalar AdoptOpenJDK? En un principio, no
Que AdoptOpenJDK mejore su soporte para GNU/Linux es algo de agradecer, sin embargo, ¿merece la pena instalarlo? En un principio no, y voy a explicar el por qué.
OpenJDK puede ser obtenido desde cualquier distribución GNU/Linux por defecto y sin añadir repositorios adicionales, ya que el sistema Open Source ha sido tradicionalmente el gran bastión de la implementación software libre de Java. Esto relega a AdoptOpenJDK a entornos de producción reales en los que se maneje distintos sistemas operativos pero buscando un entorno de producción de Java homogéneo, como por ejemplo tener oficinas en los que se encuentran Windows, Mac y GNU/Linux pero usando las mismas versiones de AdoptOpenJDK y NetBeans. Dicho con otras palabras, estos repositorios son, sobre todo, una solución para desarrolladores profesionales.
En caso de tener un entorno de producción casero o simplemente “juguetear” con Java, la utilización de AdoptOpenJDK en GNU/Linux se convierte más en una ejercicio de curiosidad que en una necesidad.
Fuente: MuyLinux
0 notes
Text



What could possibly stop us now?
#mary & george#mary and george#maryandgeorgeedit#perioddramaedit#nicholas galitzine#m&gedit#userninz#chrissiewatts#usersteen#usernuria#userclara#usermegsb#mine*#i needed to get this out my system#the cut to him and robert carr after this..... pure cinema ✨#there's some iconic lines in this show already#i cannot wait#the serve this will have. we aren't readyyyyy
344 notes
·
View notes
Text
Programming in R
Programming in R
From: http://manuals.bioinformatics.ucr.edu/home/programming-in-r
Author:
Thomas Girke
,
UC Riverside
Contents
1 Introduction
2 R Basics
3 Code Editors for R
4 Integrating R with Vim and Tmux
5 Finding Help
6 Control Structures
7 Functions
8 Useful Utilities
9 Running R Programs
10 Object-Oriented Programming (OOP)
11 Building R Packages
12 Reproducible Research by Integrating R with Latex or Markdown
13 R Programming Exercises
14 Translation of this Page
6.1 Conditional Executions
6.2 Loops
6.1.1 Comparison Operators
6.1.2 Logical Operators
6.1.3 If Statements
6.1.4 Ifelse Statements
6.2.1 For Loop
6.2.2 While Loop
6.2.3 Apply Loop Family
6.2.4 Other Loops
6.2.5 Improving Speed Performance of Loops
6.2.3.1 For Two-Dimensional Data Sets: apply
6.2.3.2 For Ragged Arrays: tapply
6.2.3.3 For Vectors and Lists: lapply and sapply
8.1 Debugging Utilities
8.2 Regular Expressions
8.3 Interpreting Character String as Expression
8.4 Time, Date and Sleep
8.5 Calling External Software with System Command
8.6 Miscellaneous Utilities
10.1 Define S4 Classes
10.2 Assign Generics and Methods
13.1 Exercise Slides
13.2 Sample Scripts
13.2.1 Batch Operations on Many Files
13.2.2 Large-scale Array Analysis
13.2.3 Graphical Procedures: Feature Map Example
13.2.4 Sequence Analysis Utilities
13.2.5 Pattern Matching and Positional Parsing of Sequences
13.2.6 Identify Over-Represented Strings in Sequence Sets
13.2.7 Translate DNA into Protein
13.2.8 Subsetting of Structure Definition Files (SDF)
13.2.9 Managing Latex BibTeX Databases
13.2.10 Loan Payments and Amortization Tables
13.2.11 Course Assignment: GC Content, Reverse & Complement
Introduction
[
Slides
] [
R Code
]
General Overview
One of the main attractions of using the
R (http://cran.at.r-project.org)
environment is the ease with which users can write their own programs and custom functions. The R programming syntax is extremely easy to learn, even for users with no previous programming experience. Once the basic R programming control structures are understood, users can use the R language as a powerful environment to perform complex custom analyses of almost any type of data.
Format of this Manual
In this manual all commands are given in code boxes, where the R code is printed in black, the comment text in blue and the output generated by R in green. All comments/explanations start with the standard comment sign '#' to prevent them from being interpreted by R as commands. This way the content in the code boxes can be pasted with their comment text into the R console to evaluate their utility. Occasionally, several commands are printed on one line and separated by a semicolon ';'. Commands starting with a '$' sign need to be executed from a Unix or Linux shell. Windows users can simply ignore them.
R Basics
The
R & BioConductor
manual provides a general introduction to the usage of the R environment and its basic command syntax.
Code Editors for R
Several excellent code editors are available that provide functionalities like R syntax highlighting, auto code indenting and utilities to send code/functions to the R console.
Basic code editors provided by Rguis
RStudio: GUI-based IDE for R
Vim-R-Tmux: R working environment based on vim and tmux
Emacs (ESS add-on package)
gedit and Rgedit
RKWard
Eclipse
Tinn-R
Notepad++ (NppToR)
Programming in R using Vim or Emacs Programming in R using RStudio
Integrating R with Vim and Tmux
Users interested in integrating R with vim and tmux may want to consult the Vim-R-Tmux configuration page.
Finding Help
Reference list on R programming (selection)
R Programming for Bioinformatics, by Robert Gentleman
Advanced R, by Hadley Wickham
S Programming, by W. N. Venables and B. D. Ripley
Programming with Data, by John M. Chambers
R Help & R Coding Conventions, Henrik Bengtsson, Lund University
Programming in R (Vincent Zoonekynd)
Peter's R Programming Pages, University of Warwick
Rtips, Paul Johnsson, University of Kansas
R for Programmers, Norm Matloff, UC Davis
High-Performance R, Dirk Eddelbuettel tutorial presented at useR-2008
C/C++ level programming for R, Gopi Goswami
Control Structures
Conditional Executions
Comparison Operators
equal: ==
not equal: !=
greater/less than: > <
greater/less than or equal: >= <=
Logical Operators
and: &
or: |
not: !
If StatementsIf statements operate on length-one logical vectors.
Syntax
if(cond1=true) { cmd1 } else { cmd2 }
Example
if(1==0) {
print(1)
} else {
print(2)
}
[1] 2
Table of Contents
Avoid inserting newlines between '} else'.
Ifelse StatementsIfelse statements operate on vectors of variable length.
Syntax
ifelse(test, true_value, false_value)
Example
x <- 1:10 # Creates sample data
ifelse(x<5 | x>8, x, 0)
[1] 1 2 3 4 0 0 0 0 9 10
Table of Contents
Loops
The most commonly used loop structures in R are for, while and apply loops. Less common are repeat loops. The break function is used to break out of loops, and next halts the processing of the current iteration and advances the looping index.
For LoopFor loops are controlled by a looping vector. In every iteration of the loop one value in the looping vector is assigned to a variable that can be used in the statements of the body of the loop. Usually, the number of loop iterations is defined by the number of values stored in the looping vector and they are processed in the same order as they are stored in the looping vector.
Syntax
for(variable in sequence) {
statements
}
Example
mydf <- iris
myve <- NULL # Creates empty storage container
for(i in seq(along=mydf[,1])) {
myve <- c(myve, mean(as.numeric(mydf[i, 1:3]))) # Note: inject approach is much faster than append with 'c'. See below for details.
}
myve
[1] 3.333333 3.100000 3.066667 3.066667 3.333333 3.666667 3.133333 3.300000
[9] 2.900000 3.166667 3.533333 3.266667 3.066667 2.800000 3.666667 3.866667
Table of Contents
Example:
condition*
x <- 1:10
z <- NULL
for(i in seq(along=x)) {
if(x[i] < 5) {
z <- c(z, x[i] - 1)
} else {
z <- c(z, x[i] / x[i])
}
}
z
[1] 0 1 2 3 1 1 1 1 1 1
Table of Contents
Example:
stop on condition and print error message
x <- 1:10
z <- NULL
for(i in seq(along=x)) {
if (x[i]<5) {
z <- c(z,x[i]-1)
} else {
stop("values need to be <5")
}
}
Error: values need to be <5
z
[1] 0 1 2 3
Table of Contents
While LoopSimilar to for loop, but the iterations are controlled by a conditional statement.
Syntax
while(condition) statements
Example
z <- 0
while(z < 5) {
z <- z + 2
print(z)
}
[1] 2
[1] 4
[1] 6
Table of Contents
Apply Loop Family
For T
wo-Dimensional Data Sets: apply
Syntax
apply(X, MARGIN, FUN, ARGs)
X: array, matrix or data.frame; MARGIN: 1 for rows, 2 for columns, c(1,2) for both; FUN: one or more functions; ARGs: possible arguments for function
Example
## Example for applying predefined mean function
apply(iris[,1:3], 1, mean)
[1] 3.333333 3.100000 3.066667 3.066667 3.333333 3.666667 3.133333 3.300000
...
## With custom function
x <- 1:10
test <- function(x) {
# Defines some custom function
if(x < 5) {
x-1
} else {
x / x
}
}
apply(as.matrix(x), 1, test) #
Returns same result as previous for loop*
[1] 0 1 2 3 1 1 1 1 1 1
## Same as above but with a single line of code
apply(as.matrix(x), 1, function(x) { if (x<5) { x-1 } else { x/x } })
[1] 0 1 2 3 1 1 1 1 1 1
Table of Contents
For Ragged Arrays: tapply
Applies a function to array categories of variable lengths (ragged array). Grouping is defined by factor.
Syntax
tapply(vector, factor, FUN)
Example
## Computes mean values of vector agregates defined by factor
tapply(as.vector(iris[,4]), factor(iris[,5]), mean)
setosa versicolor virginica
0.246 1.326 2.026
## The aggregate function provides related utilities
aggregate(iris[,1:4], list(iris$Species), mean)
Group.1 Sepal.Length Sepal.Width Petal.Length Petal.Width
1 setosa 5.006 3.428 1.462 0.246
2 versicolor 5.936 2.770 4.260 1.326
3 virginica 6.588 2.974 5.552 2.026
Table of Contents
For Vectors and Lists: lapply and sapply
Both apply a function to vector or list objects. The function lapply returns a list, while sapply attempts to return the simplest data object, such as vector or matrix instead of list.
Syntax
lapply(X, FUN)
sapply(X, FUN)
Example
## Creates a sample list
mylist <- as.list(iris[1:3,1:3])
mylist
$Sepal.Length
[1] 5.1 4.9 4.7
$Sepal.Width
[1] 3.5 3.0 3.2
$Petal.Length
[1] 1.4 1.4 1.3
## Compute sum of each list component and return result as list
lapply(mylist, sum)
$Sepal.Length
[1] 14.7
$Sepal.Width
[1] 9.7
$Petal.Length
[1] 4.1
##
Compute sum of each list component and return result as vector
sapply(mylist, sum)
Sepal.Length Sepal.Width Petal.Length
14.7 9.7 4.1
Table of Contents
Other Loops
Repeat Loop
Syntax
repeat statement
s
Loop is repeated until a break is specified. This means there needs to be a second statement to test whether or not to break from the loop.
Example
z <- 0
repeat {
z <- z + 1
print(z)
if(z > 100) break()
}
Table of Contents
Improving Speed Performance of LoopsLooping over very large data sets can become slow in R. However, this limitation can be overcome by eliminating certain operations in loops or avoiding loops over the data intensive dimension in an object altogether. The latter can be achieved by performing mainly vector-to-vecor or matrix-to-matrix computations which run often over 100 times faster than the corresponding for() or apply() loops in R. For this purpose, one can make use of the existing speed-optimized R functions (
e.g.:
rowSums, rowMeans, table, tabulate) or one can design custom functions that avoid expensive R loops by using vector- or matrix-based approaches. Alternatively, one can write programs that will perform all time consuming computations on the C-level.
(1)
Speed comparison of for loops with an append versus an inject step:
myMA <- matrix(rnorm(1000000), 100000, 10, dimnames=list(1:100000, paste("C", 1:10, sep="")))
results <- NULL
system.time(for(i in seq(along=myMA[,1])) results <- c(results, mean(myMA[i,])))
user system elapsed
39.156 6.369 45.559
results <- numeric(length(myMA[,1]))
system.time(for(i in seq(along=myMA[,1])) results[i] <- mean(myMA[i,]))
user system elapsed
1.550 0.005 1.556
Table of Contents
The inject approach is 20-50 times faster than the append version.
(2)
Speed comparison of apply loop versus rowMeans for computing the mean for each row in a large matrix:
system.time(myMAmean <- apply(myMA, 1, mean))
user system elapsed
1.452 0.005 1.456
system.time(myMAmean <- rowMeans(myMA))
user system elapsed
0.005 0.001 0.006
Table of Contents
The rowMeans approach is over 200 times faster than the apply loop.
(3)
Speed comparison of apply loop versus vectorized approach for computing the standard deviation of each row:
system.time(myMAsd <- apply(myMA, 1, sd))
user system elapsed
3.707 0.014 3.721
myMAsd[1:4]
1 2 3 4
0.8505795 1.3419460 1.3768646 1.3005428
system.time(myMAsd <- sqrt((rowSums((myMA-rowMeans(myMA))^2)) / (length(myMA[1,])-1)))
user system elapsed
0.020 0.009 0.028
myMAsd[1:4]
1 2 3 4
0.8505795 1.3419460 1.3768646 1.3005428
Table of Contents
The vector-based approach in the last step is over 200 times faster than the apply loop.
(4)
Example for computing the mean for any custom selection of columns without compromising the speed performance:
## In the following the colums are named according to their selection in myList
myList <- tapply(colnames(myMA), c(1,1,1,2,2,2,3,3,4,4), list)
myMAmean <- sapply(myList, function(x) rowMeans(myMA[,x]))
colnames(myMAmean) <- sapply(myList, paste, collapse="_")
myMAmean[1:4,]
C1_C2_C3 C4_C5_C6 C7_C8 C9_C10
1 0.0676799 -0.2860392 0.09651984 -0.7898946
2 -0.6120203 -0.7185961 0.91621371 1.1778427
3 0.2960446 -0.2454476 -1.18768621 0.9019590
4 0.9733695 -0.6242547 0.95078869 -0.7245792
## Alternative to achieve the same result with similar performance, but in a much less elegant way
myselect <- c(1,1,1,2,2,2,3,3,4,4) # The colums are named according to the selection stored in myselect
myList <- tapply(seq(along=myMA[1,]), myselect, function(x) paste("myMA[ ,", x, "]", sep=""))
myList <- sapply(myList, function(x) paste("(", paste(x, collapse=" + "),")/", length(x)))
myMAmean <- sapply(myList, function(x) eval(parse(text=x)))
colnames(myMAmean) <- tapply(colnames(myMA), myselect, paste, collapse="_")
myMAmean[1:4,]
C1_C2_C3 C4_C5_C6 C7_C8 C9_C10
1 0.0676799 -0.2860392 0.09651984 -0.7898946
2 -0.6120203 -0.7185961 0.91621371 1.1778427
3 0.2960446 -0.2454476 -1.18768621 0.9019590
4 0.9733695 -0.6242547 0.95078869 -0.7245792
Table of Contents
Functions
A very useful feature of the R environment is the possibility to expand existing functions and to easily write custom functions. In fact, most of the R software can be viewed as a series of R functions.
Syntax to define functions
myfct <- function(arg1, arg2, ...) {
function_body
}
Table of Contents
The value returned by a function is the value of the function body, which is usually an unassigned final expression,
e.g.:
return()
Syntax to call functions
myfct(arg1=..., arg2=...)
Table of Contents
Syntax Rules for Functions
General
Functions are defined by (1) assignment with the keyword function, (2) the declaration of arguments/variables (arg1, arg2, ...) and (3) the definition of operations (function_body) that perform computations on the provided arguments. A function name needs to be assigned to call the function (see below).
Naming
Function names can be almost anything. However, the usage of names of existing functions should be avoided.
Arguments
It is often useful to provide default values for arguments (
e.g.
:arg1=1:10). This way they don't need to be provided in a function call. The argument list can also be left empty (myfct <- function() { fct_body }) when a function is expected to return always the same value(s). The argument '...' can be used to allow one function to pass on argument settings to another.
Function body
The actual expressions (commands/operations) are defined in the function body which should be enclosed by braces. The individual commands are separated by semicolons or new lines (preferred).
Calling functions
Functions are called by their name followed by parentheses containing possible argument names. Empty parenthesis after the function name will result in an error message when a function requires certain arguments to be provided by the user. The function name alone will print the definition of a function.
Scope
Variables created inside a function exist only for the life time of a function. Thus, they are not accessible outside of the function. To force variables in functions to exist globally, one can use this special assignment operator: '<<-'. If a global variable is used in a function, then the global variable will be masked only within the function.
Example:
Function basics
myfct <- function(x1, x2=5) {
z1 <- x1/x1
z2 <- x2*x2
myvec <- c(z1, z2)
return(myvec)
}
myfct # prints definition of function
myfct(x1=2, x2=5) # applies function to values 2 and 5
[1] 1 25
myfct(2, 5) # the argument names are not necessary, but then the order of the specified values becomes important
myfct(x1=2) # does the same as before, but the default value '5' is used in this case
Table of Contents
Example:
Function with optional arguments
myfct2 <- function(x1=5, opt_arg) {
if(missing(opt_arg)) { # 'missing()' is used to test whether a value was specified as an argument
z1 <- 1:10
} else {
z1 <- opt_arg
}
cat("my function returns:", "\n")
return(z1/x1)
}
myfct2(x1=5) # performs calculation on default vector (z1) that is defined in the function body
my function returns:
[1] 0.2 0.4 0.6 0.8 1.0 1.2 1.4 1.6 1.8 2.0
myfct2(x1=5, opt_arg=30:20) # a custom vector is used instead when the optional argument (opt_arg) is specified
my function returns:
[1] 6.0 5.8 5.6 5.4 5.2 5.0 4.8 4.6 4.4 4.2 4.0
Table of Contents
Control utilities for functions: return, warning and stop
Return
The evaluation flow of a function may be terminated at any stage with the return function. This is often used in combination with conditional evaluations.
Stop
To stop the action of a function and print an error message, one can use the stop function.
Warning
To print a warning message in unexpected situations without aborting the evaluation flow of a function, one can use the function warning("...").
myfct <- function(x1) {
if (x1>=0) print(x1) else stop("This function did not finish, because x1 < 0")
warning("Value needs to be > 0")
}
myfct(x1=2)
[1] 2
Warning message:
In myfct(x1 = 2) : Value needs to be > 0
myfct(x1=-2)
Error in myfct(x1 = -2) : This function did not finish, because x1 < 0
Table of Contents
Useful Utilities
Debugging Utilities
Several debugging utilities are available for R. The most important utilities are: traceback(), browser(), options(error=recover), options(error=NULL) and debug(). The
Debugging in R
page provides an overview of the available resources.
Regular Expressions
R's regular expression utilities work similar as in other languages. To learn how to use them in R, one can consult the main help page on this topic with ?regexp. The following gives a few basic examples.
The grep function can be used for finding patterns in strings, here letter A in vector month.name.
month.name[grep("A", month.name)]
[1] "April" "August"
Table of Contents
Example for using regular expressions to substitute a pattern by another one using the sub/gsub function with a back reference. Remember: single escapes '\' need to be double escaped '\\' in R.
gsub("(i.*a)", "xxx_\\1", "virginica", perl = TRUE)
[1] "vxxx_irginica"
Table of Contents
Example for split and paste functions
x <- gsub("(a)", "\\1_", month.name[1], perl=TRUE) # performs substitution with back reference which inserts in this example a '_' character
x
[1] "Ja_nua_ry"
strsplit(x, "_") # splits string on inserted character from above
[[1]]
[1] "Ja" "nua" "ry"
paste(rev(unlist(strsplit(x, NULL))), collapse="") # reverses character string by splitting first all characters into vector fields and then collapsing them with paste
[1] "yr_aun_aJ"
Table of Contents
Example for importing specific lines in a file with a regular expression. The following example demonstrates the retrieval of specific lines from an external file with a regular expression. First, an external file is created with the cat function, all lines of this file are imported into a vector with readLines, the specific elements (lines) are then retieved with the grep function, and the resulting lines are split into vector fields with strsplit.
cat(month.name, file="zzz.txt", sep="\n")
x <- readLines("zzz.txt")
x <- x[c(grep("^J", as.character(x), perl = TRUE))]
t(as.data.frame(strsplit(x, "u")))
[,1] [,2]
c..Jan....ary.. "Jan" "ary"
c..J....ne.. "J" "ne"
c..J....ly.. "J" "ly"
Table of Contents
Interpreting Character String as Expression
Example
mylist <- ls() # generates vector of object names in session
mylist[1] # prints name of 1st entry in vector but does not execute it as expression that returns values of 10th object
get(mylist[1]) # uses 1st entry name in vector and executes it as expression
eval(parse(text=mylist[1])) # alternative approach to obtain similar result
Table of Contents
Time, Date and Sleep
Example
system.time(ls()) # returns CPU (and other) times that an expression used, here ls()
user system elapsed
0 0 0
date() # returns the current system date and time
[1] "Wed Dec 11 15:31:17 2012"
Sys.sleep(1) # pause execution of R expressions for a given number of seconds (e.g. in loop)
Table of Contents
Calling External Software with System CommandThe system command allows to call any command-line software from within R on Linux, UNIX and OSX systems.
system("...") # provide under '...' command to run external software e.g. Perl, Python, C++ programs
Table of Contents
Related utilities on Windows operating systems
x <- shell("dir", intern=T) # reads current working directory and assigns to file
shell.exec("C:/Documents and Settings/Administrator/Desktop/my_file.txt") # opens file with associated program
Table of Contents
Miscellaneous Utilities
(1)
Batch import and export of many files.
In the following example all file names ending with *.txt in the current directory are first assigned to a list (the '$' sign is used to anchor the match to the end of a string). Second, the files are imported one-by-one using a for loop where the original names are assigned to the generated data frames with the assign function. Consult help with ?read.table to understand arguments row.names=1 and comment.char = "A". Third, the data frames are exported using their names for file naming and appending the extension *.out.
files <- list.files(pattern=".txt$")
for(i in files) {
x <- read.table(i, header=TRUE, comment.char = "A", sep="\t")
assign(i, x)
print(i)
write.table(x, paste(i, c(".out"), sep=""), quote=FALSE, sep="\t", col.names = NA)
}
Table of Contents
(2)
Running Web Applications (basics on designing web client/crawling/scraping scripts in R)
Example for obtaining MW values for peptide sequences from the
EXPASY's pI/MW Tool
web page.
myentries <- c("MKWVTFISLLFLFSSAYS", "MWVTFISLL", "MFISLLFLFSSAYS")
myresult <- NULL
for(i in myentries) {
myurl <- paste("http://ca.expasy.org/cgi-bin/pi_tool?protein=", i, "&resolution=monoisotopic", sep="")
x <- url(myurl)
res <- readLines(x)
close(x)
mylines <- res[grep("Theoretical pI/Mw:", res)]
myresult <- c(myresult, as.numeric(gsub(".*/ ", "", mylines)))
print(myresult)
Sys.sleep(1) # halts process for one sec to give web service a break
}
final <- data.frame(Pep=myentries, MW=myresult)
cat("\n The MW values for my peptides are:\n")
final
Pep MW
1 MKWVTFISLLFLFSSAYS 2139.11
2 MWVTFISLL 1108.60
3 MFISLLFLFSSAYS 1624.82
Table of Content
Running R Programs
(1)
Executing an R script from the R console
source("my_script.R")
Table of Contents
(2.1)
Syntax for running R programs from the command-line. Requires in first line of my_script.R the following statement: #!/usr/bin/env Rscript
$ Rscript my_script.R # or just ./myscript.R after making file executable with 'chmod +x my_script.R'
All commands starting with a '$' sign need to be executed from a Unix or Linux shell.
(2.2)
Alternatively, one can use the following syntax to run R programs in BATCH mode from the command-line.
$ R CMD BATCH [options] my_script.R [outfile]
The output file lists the commands from the script file and their outputs. If no outfile is specified, the name used is that of infile and .Rout is appended to outfile. To stop all the usual R command line information from being written to the outfile, add this as first line to my_script.R file: options(echo=FALSE). If the command is run like this R CMD BATCH --no-save my_script.R, then nothing will be saved in the .Rdata file which can get often very large. More on this can be found on the help pages: $ R CMD BATCH --help or ?BATCH.
(2.3)
Another alternative for running R programs as silently as possible.
$ R --slave < my_infile > my_outfile
Argument --slave makes R run as 'quietly' as possible.
(3)
Passing Command-Line Arguments to R Programs
Create an R script, here named test.R, like this one:
######################
myarg <- commandArgs()
print(iris[1:myarg[6], ])
######################
Then run it from the command-line like this:
$ Rscript test.R 10
In the given example the number 10 is passed on from the command-line as an argument to the R script which is used to return to STDOUT the first 10 rows of the iris sample data. If several arguments are provided, they will be interpreted as one string that needs to be split it in R with the strsplit function.
(4)
Submitting R script to a Linux cluster via Torque
Create the following shell script my_script.sh
#################################
#!/bin/bash
cd $PBS_O_WORKDIR
R CMD BATCH --no-save my_script.R
#################################
This script doesn't need to have executable permissions. Use the following qsub command to send this shell script to the Linux cluster from the directory where the R script my_script.R is located. To utilize several CPUs on the Linux cluster, one can divide the input data into several smaller subsets and execute for each subset a separate process from a dedicated directory.
$ qsub my_script.sh
Table of Contents
Here is a short R script that generates the required files and directories automatically and submits the jobs to the nodes:
submit2cluster.R
. For more details, see also this
'Tutorial on Parallel Programming in R' by Hanna Sevcikova
(5)
Submitting jobs to Torque or any other queuing/scheduling system via the
BatchJobs
package. This package provides one of the most advanced resources for submitting jobs to queuing systems from within R. A related package is
BiocParallel
from Bioconductor which extends many functionalities of BatchJobs to genome data analysis. Useful documentation for BatchJobs:
Technical Report
,
GitHub page
,
Slide Show
,
Config samples
.
library(BatchJobs)
loadConfig(conffile = ".BatchJobs.R")
## Loads configuration file. Here .BatchJobs.R containing just this line:
## cluster.functions <- makeClusterFunctionsTorque("torque.tmpl")
## The template file torque.tmpl is expected to be in the current working
## director. It can be downloaded from here:
## https://github.com/tudo-r/BatchJobs/blob/master/examples/cfTorque/simple.tmpl
getConfig() # Returns BatchJobs configuration settings
reg <- makeRegistry(id="BatchJobTest", work.dir="results")
## Constructs a registry object. Output files from R will be stored under directory "results",
## while the
standard objects from BatchJobs will be stored in the directory "BatchJobTest-files".
print(reg)
## Some test function
f <- function(x) {
system("ls -al >> test.txt")
x
}
## Adds jobs to registry object (here reg)
ids <- batchMap(reg, fun=f, 1:10)
print(ids)
showStatus(reg)
## Submit jobs or chunks of jobs to batch system via cluster function
done <- submitJobs(reg, resources=list(walltime=3600, nodes="1:ppn=4", memory="4gb"))
## Load results from BatchJobTest-files/jobs/01/1-result.RData
loadResult(reg, 1)
Table of Contents
Object-Oriented Programming (OOP)
R supports two systems for object-oriented programming (OOP). An older S3 system and a more recently introduced S4 system. The latter is more formal, supports multiple inheritance, multiple dispatch and introspection. Many of these features are not available in the older S3 system. In general, the OOP approach taken by R is to separate the class specifications from the specifications of generic functions (function-centric system). The following introduction is restricted to the S4 system since it is nowadays the preferred OOP method for R. More information about OOP in R can be found in the following introductions:
Vincent Zoonekynd's introduction to S3 Classes
,
S4 Classes in 15 pages
,
Christophe Genolini's S4 Intro
,
The R.oo package
,
BioC Course: Advanced R for Bioinformatics
,
Programming with R by John Chambers
and
R Programming for Bioinformatics by Robert Gentleman
.
Define S4 Classes
(A)
Define S4 Classes with
setClass() and new()
y <- matrix(1:50, 10, 5) # Sample data set
setClass(Class="myclass",
representation=representation(a="ANY"),
prototype=prototype(a=y[1:2,]), # Defines default value (optional)
validity=function(object) { # Can be defined in a separate step using setValidity
if(class(object@a)!="matrix") {
return(paste("expected matrix, but obtained", class(object@a)))
} else {
return(TRUE)
}
}
)
Table of Contents
The setClass function defines classes. Its most important arguments are
Class: the name of the class
representation: the slots that the new class should have and/or other classes that this class extends.
prototype: an object providing default data for the slots.
contains: the classes that this class extends.
validity, access, version: control arguments included for compatibility with S-Plus.
where: the environment to use to store or remove the definition as meta data.
(B)
The function new creates an instance of a class (here myclass)
myobj <- new("myclass", a=y)
myobj
An object of class "myclass"
Slot "a":
[,1] [,2] [,3] [,4] [,5]
[1,] 1 11 21 31 41
[2,] 2 12 22 32 42
...
new("myclass", a=iris) # Returns an error message due to wrong input type (iris is data frame)
Error in validObject(.Object) :
invalid class “myclass” object: expected matrix, but obtained data.frame
Table of Contents
Its arguments are:
Class: the name of the class
...: Data to include in the new object with arguments according to slots in class definition.
(C)
A more generic way of creating class instances is to define an initialization method (details below)
setMethod("initialize", "myclass", function(.Object, a) {
.Object@a <- a/a
.Object
})
new("myclass", a = y)
[1] "initialize"
new("myclass", a = y)> new("myclass", a = y)
An object of class "myclass"
Slot "a":
[,1] [,2] [,3] [,4] [,5]
[1,] 1 1 1 1 1
[2,] 1 1 1 1 1
...
Table of Contents
(D)
Usage and helper functions
myobj@a # The '@' extracts the contents of a slot. Usage should be limited to internal functions!
initialize(.Object=myobj, a=as.matrix(cars[1:3,])) # Creates a new S4 object from an old one.
# removeClass("myclass") # Removes object from current session; does not apply to associated methods.
Table of Contents
(E)
Inheritance: allows to define new classes that inherit all properties (
e.g.
data slots, methods) from their existing parent classes
setClass("myclass1", representation(a = "character", b = "character"))
setClass("myclass2", representation(c = "numeric", d = "numeric"))
setClass("myclass3", contains=c("myclass1", "myclass2"))
new("myclass3", a=letters[1:4], b=letters[1:4], c=1:4, d=4:1)
An object of class "myclass3"
Slot "a":
[1] "a" "b" "c" "d"
Slot "b":
[1] "a" "b" "c" "d"
Slot "c":
[1] 1 2 3 4
Slot "d":
[1] 4 3 2 1
getClass("myclass1")
Class "myclass1" [in ".GlobalEnv"]
Slots:
Name: a b
Class: character character
Known Subclasses: "myclass3"
getClass("myclass2")
Class "myclass2" [in ".GlobalEnv"]
Slots:
Name: c d
Class: numeric numeric
Known Subclasses: "myclass3"
getClass("myclass3")
Class "myclass3" [in ".GlobalEnv"]
Slots:
Name: a b c d
Class: character character numeric numeric
Extends: "myclass1", "myclass2"
Table of Contents
The argument contains allows to extend existing classes; this propagates all slots of parent classes.
(F)
Coerce objects to another class
setAs(from="myclass", to="character", def=function(from) as.character(as.matrix(from@a)))
as(myobj, "character")
[1] "1" "2" "3" "4" "5" "6" "7" "8" "9" "10" "11" "12" "13" "14" "15"
...
Table of Contents
(G)
Virtual classes are constructs for which no instances will be or can be created. They are used to link together classes which may have distinct representations (e.g. cannot inherit from each other) but for which one wants to provide similar functionality. Often it is desired to create a virtual class and to then have several other classes extend it. Virtual classes can be defined by leaving out the representation argument or including the class VIRTUAL:
setClass("myVclass")
setClass("myVclass", representation(a = "character", "VIRTUAL"))
Table of Contents
(H)
Functions to introspect classes
getClass("myclass")
getSlots("myclass")
slotNames("myclass")
extends("myclass2")
Assign Generics and MethodsAssign generics and methods with setGeneric() and setMethod()
(A)
Accessor function (to avoid usage of '@')
setGeneric(name="acc", def=function(x) standardGeneric("acc"))
setMethod(f="acc", signature="myclass", definition=function(x) {
return(x@a)
})
acc(myobj)
[,1] [,2] [,3] [,4] [,5]
[1,] 1 11 21 31 41
[2,] 2 12 22 32 42
...
Table of Contents
(B.1)
Replacement method using custom accessor function (acc <-)
setGeneric(name="acc<-", def=function(x, value) standardGeneric("acc<-"))
setReplaceMethod(f="acc", signature="myclass", definition=function(x, value) {
x@a <- value
return(x)
})
## After this the following replace operations with 'acc' work on new object class
acc(myobj)[1,1] <- 999 # Replaces first value
colnames(acc(myobj)) <- letters[1:5] # Assigns new column names
rownames(acc(myobj)) <- letters[1:10] # Assigns new row names
myobj
An object of class "myclass"
Slot "a":
a b c d e
a 999 11 21 31 41
b 2 12 22 32 42
...
Table of Contents
(B.2)
Replacement method using "[" operator ([<-)
setReplaceMethod(f="[", signature="myclass", definition=function(x, i, j, value) {
x@a[i,j] <- value
return(x)
})
myobj[1,2] <- 999
myobj
An object of class "myclass"
Slot "a":
a b c d e
a 999 999 21 31 41
b 2 12 22 32 42
...
Table of Contents
(C)
Define behavior of "[" subsetting operator (no generic required!)
setMethod(f="[", signature="myclass",
definition=function(x, i, j, ..., drop) {
x@a <- x@a[i,j]
return(x)
})
myobj[1:2,] # Standard subsetting works now on new class
An object of class "myclass"
Slot "a":
a b c d e
a 999 999 21 31 41
b 2 12 22 32 42
...
Table of Contents
(D)
Define print behavior
setMethod(f= show", signature="myclass", definition=function(object) {
cat("An instance of ", "\"", class(object), "\"", " with ", length(acc(object)[,1]), " elements", "\n", sep="")
if(length(acc(object)[,1])>=5) {
print(as.data.frame(rbind(acc(object)[1:2,], ...=rep("...", length(acc(object)[1,])),
acc(object)[(length(acc(object)[,1])-1):length(acc(object)[,1]),])))
} else {
print(acc(object))
}})
myobj # Prints object with custom method
An instance of "myclass" with 10 elements
a b c d e
a 999 999 21 31 41
b 2 12 22 32 42
... ... ... ... ... ...
i 9 19 29 39 49
j 10 20 30 40 50
Table of Contents
(E)
Define a data specific function (here randomize row order)
setGeneric(name="randomize", def=function(x) standardGeneric("randomize"))
setMethod(f="randomize", signature="myclass", definition=function(x) {
acc(x)[sample(1:length(acc(x)[,1]), length(acc(x)[,1])), ]
})
randomize(myobj)
a b c d e
j 10 20 30 40 50
b 2 12 22 32 42
...
Table of Contents
(F)
Define a graphical plotting function and allow user to access it with generic plot function
setMethod(f="plot", signature="myclass", definition=function(x, ...) {
barplot(as.matrix(acc(x)), ...)
})
plot(myobj)
Table of Contents
(G)
Functions to inspect methods
showMethods(class="myclass")
findMethods("randomize")
getMethod("randomize", signature="myclass")
existsMethod("randomize", signature="myclass")
Building R Packages
To get familiar with the structure, building and submission process of R packages, users should carefully read the documentation on this topic available on these sites:
Writing R Extensions, R web site
R Packages, by Hadley Wickham
R Package Primer, by Karl Broman
Package Guidelines, Bioconductor
Advanced R Programming Class, Bioconductor
Short Overview of Package Building Process
(A)
Automatic package building with the package.skeleton function:
package.skeleton(name="mypackage", code_files=c("script1.R", "script2.R"))
Table of Contents
Note: this is an optional but very convenient function to get started with a new package. The given example will create a directory named mypackage containing the skeleton of the package for all functions, methods and classes defined in the R script(s) passed on to the code_files argument. The basic structure of the package directory is
described here
. The package directory will also contain a file named 'Read-and-delete-me' with the following instructions for completing the package:
Edit the help file skeletons in man, possibly combining help files for multiple functions.
Edit the exports in NAMESPACE, and add necessary imports.
Put any C/C++/Fortran code in src.
If you have compiled code, add a useDynLib() directive to NAMESPACE.
Run R CMD build to build the package tarball.
Run R CMD check to check the package tarball.
Read Writing R Extensions for more information.
(B)
Once a package skeleton is available one can build the package from the command-line (Linux/OS X):
$ R CMD build mypackage
Table of Contents
This will create a tarball of the package with its version number encoded in the file name, e.g.: mypackage_1.0.tar.gz.
Subsequently, the package tarball needs to be checked for errors with:
$ R CMD check mypackage_1.0.tar.gz
Table of Contents
All issues in a package's source code and documentation should be addressed until R CMD check returns no error or warning messages anymore.
(C)
Install package from source:
Linux:
install.packages("mypackage_1.0.tar.gz", repos=NULL)
Table of Contents
OS X:
install.packages("mypackage_1.0.tar.gz", repos=NULL, type="source")
Table of Contents
Windows requires a zip archive for installing R packages, which can be most conveniently created from the command-line (Linux/OS X) by installing the package in a local directory (here tempdir) and then creating a zip archive from the installed package directory:
$ mkdir tempdir
$ R CMD INSTALL -l tempdir mypackage_1.0.tar.gz
$ cd tempdir
$ zip -r mypackage mypackage
## The resulting mypackage.zip archive can be installed under Windows like this:
install.packages("mypackage.zip", repos=NULL)
Table of Contents
This procedure only works for packages which do not rely on compiled code (C/C++). Instructions to fully build an R package under Windows can be
found here
and
here
.
(D)
Maintain/expand an existing package:
Add new functions, methods and classes to the script files in the ./R directory in your package
Add their names to the NAMESPACE file of the package
Additional *.Rd help templates can be generated with the prompt*() functions like this:
source("myscript.R") # imports functions, methods and classes from myscript.R
prompt(myfct) # writes help file myfct.Rd
promptClass("myclass") # writes file myclass-class.Rd
promptMethods("mymeth") # writes help file mymeth.Rd
Table of Contents
The resulting *.Rd help files can be edited in a text editor and properly rendered and viewed from within R like this:
library(tools)
Rd2txt("./mypackage/man/myfct.Rd") # renders *.Rd files as they look in final help pages
checkRd("./mypackage/man/myfct.Rd") # checks *.Rd help file for problems
Table of Contents
(E)
Submit package to a public repository
The best way of sharing an R package with the community is to submit it to one of the main R package repositories, such as CRAN or Bioconductor. The details about the submission process are given on the corresponding repository submission pages:
Submitting to Bioconductor (guidelines, submission, svn control, build/checks release, build/checks devel)
Submitting to CRAN
Reproducible Research by Integrating R with Latex or Markdown
See Sweave/Stangle sections of Slide Show for this manual
Sweave Manual
R Markdown
knitr manual
RStudio's Rpubs page
R Programming Exercises
Exercise Slides[
Slides
] [
Exercises
] [
Additional Exercises
]
Download on of the above exercise files, then start editing this R source file with a programming text editor, such as Vim, Emacs or one of the R GUI text editors. Here is the
HTML version
of the code with syntax coloring.
Sample Scripts
Batch Operations on Many Files
## (1) Start R from an empty test directory
## (2) Create some files as sample data
for(i in month.name) {
mydf <- data.frame(Month=month.name, Rain=runif(12, min=10, max=100), Evap=runif(12, min=1000, max=2000))
write.table(mydf, file=paste(i , ".infile", sep=""), quote=F, row.names=F, sep="\t")
}
## (3) Import created files, perform calculations and export to renamed files
files <- list.files(pattern=".infile$")
for(i in seq(along=files)) { # start for loop with numeric or character vector; numeric vector is often more flexible
x <- read.table(files[i], header=TRUE, row.names=1, comment.char = "A", sep="\t")
x <- data.frame(x, sum=apply(x, 1, sum), mean=apply(x, 1, mean)) # calculates sum and mean for each data frame
assign(files[i], x) # generates data frame object and names it after content in variable 'i'
print(files[i], quote=F) # prints loop iteration to screen to check its status
write.table(x, paste(files[i], c(".out"), sep=""), quote=FALSE, sep="\t", col.names = NA)
}
## (4) Same as above, but file naming by index data frame. This way one can organize file names by external table.
name_df <- data.frame(Old_name=sort(files), New_name=sort(month.abb))
for(i in seq(along=name_df[,1])) {
x <- read.table(as.vector(name_df[i,1]), header=TRUE, row.names=1, comment.char = "A", sep="\t")
x <- data.frame(x, sum=apply(x, 1, sum), mean=apply(x, 1, mean))
assign(as.vector(name_df[i,2]), x) # generates data frame object and names it after 'i' entry in column 2
print(as.vector(name_df[i,1]), quote=F)
write.table(x, paste(as.vector(name_df[i,2]), c(".out"), sep=""), quote=FALSE, sep="\t", col.names = NA)
}
## (5) Append content of all input files to one file.
files <- list.files(pattern=".infile$")
all_files <- data.frame(files=NULL, Month=NULL, Gain=NULL , Loss=NULL, sum=NULL, mean=NULL) # creates empty data frame container
for(i in seq(along=files)) {
x <- read.table(files[i], header=TRUE, row.names=1, comment.char = "A", sep="\t")
x <- data.frame(x, sum=apply(x, 1, sum), mean=apply(x, 1, mean))
x <- data.frame(file=rep(files[i], length(x[,1])), x) # adds file tracking column to x
all_files <- rbind(all_files, x) # appends data from all files to data frame 'all_files'
write.table(all_files, file="all_files.xls", quote=FALSE, sep="\t", col.names = NA)
}
## (6) Write the above code into a text file and execute it with the commands 'source' and 'BATCH'.
source("my_script.R") # execute from R console
$ R CMD BATCH my_script.R # execute from shell
Table of Contents
Large-scale Array Analysis
Sample script to perform large-scale expression array analysis with complex queries: lsArray.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/lsArray.R")
Table of Contents
Graphical Procedures: Feature Map Example
Script to plot feature maps of genes or chromosomes: featureMap.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/featureMap.txt")
Table of Contents
Sequence Analysis UtilitiesIncludes sequence batch import, sub-setting, pattern matching, AA Composition, NEEDLE, PHYLIP, etc. The script '
sequenceAnalysis.R
' demonstrates how R can be used as a powerful tool for managing and analyzing large sets of biological sequences. This example also shows how easy it is to integrate R with the
EMBOSS
project or other external programs. The script provides the following functionality:
Batch sequence import into R data frame
Motif searching with hit statistics
Analysis of sequence composition
All-against-all sequence comparisons
Generation of phylogenetic trees
To demonstrate the utilities of the script, users can simply execute it from R with the following source command:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/sequenceAnalysis.txt")
Table of Contents
Pattern Matching and Positional Parsing of Sequences
Functions for importing sequences into R, retrieving reverse and complement of nucleotide sequences, pattern searching, positional parsing and exporting search results in HTML format: patternSearch.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/patternSearch.R")
Table of Contents
Identify Over-Represented Strings in Sequence Sets
Functions for finding over-represented words in sets of DNA, RNA or protein sequences: wordFinder.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/wordFinder.R")
Table of Contents
Translate DNA into Protein
Script 'translateDNA.R' for translating NT sequences into AA sequences (required codon table). To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/translateDNA.R")
Table of Contents
Subsetting of Structure Definition Files (SDF)
Script for importing and subsetting SDF files: sdfSubset.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/sdfSubset.R")
Table of Contents
Managing Latex BibTeX Databases
Script for importing BibTeX databases into R, retrieving the individual references with a full-text search function and viewing the results in R or in HubMed: BibTex.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/BibTex.R")
Table of Contents
Loan Payments and Amortization Tables
This script calculates monthly and annual mortgage or loan payments, generates amortization tables and plots the results: mortgage.R. To demo what the script does, run it like this:
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/mortgage.R")
Table of Contents
Course Assignment: GC Content, Reverse & Complement
Apply the above information to write a function that calculates for a set of DNA sequences their GC content and generates their reverse and complement. Here are some useful commands that can be incorporated in this function:
## Generate an example data frame with ID numbers and DNA sequences
fx <- function(test) {
x <- as.integer(runif(20, min=1, max=5))
x[x==1] <- "A"; x[x==2] <- "T"; x[x==3] <- "G"; x[x==4] <- "C"
paste(x, sep = "", collapse ="")
}
z1 <- c()
for(i in 1:50) {
z1 <- c(fx(i), z1)
}
z1 <- data.frame(ID=seq(along=z1), Seq=z1)
z1
## Write each character of sequence into separate vector field and reverse its order
my_split <- strsplit(as.character(z1[1,2]),"")
my_rev <- rev(my_split[[1]])
paste(my_rev, collapse="")
## Generate the sequence complement by replacing G|C|A|T by C|G|T|A
## Use 'apply' or 'for loop' to apply the above operations to all sequences in sample data frame 'z1'
## Calculate in the same loop the GC content for each sequence using the following command
table(my_split[[1]])/length(my_split[[1]])
Table of Contents
0 notes
Text







i think it's time for a new member in your bedchamber.
MARY & GEORGE (2024) → Episode 2, The Hunt
#mary & george#george villiers#james i#nicholas galitzine#tony curran#mary and george#maryandgeorgeedit#m&gedit#nicholasgalitzineedit#userstream#dailyfilmtvgifs#perioddramacentral#swearphil#userlang#iuserzoe#chrissiewatts#*#my edit
264 notes
·
View notes
Text
Audio on Intel HDA Machines
Many Skywave Linux users with Dell, Hp, and Compaq hardware have the "no sound" issue. It stems from pulse and alsa not doing a good job of automatically finding and configuring the particular soundcards used on those computers. It is an irritating problem that has existed for years and particularly bothersome on Skywave Linux due to the need to specify sinks and plugins for the audio processing chain. If you have hardware which does not have functioning sound on initial boot of Skywave Linux, consider doing a persistent USB or full installation where system changes are not lost at shutdown. Make note of the cards and subdevices on your system. In a terminal, enter the command "aplay -l" without the quotes. Some systems have only one card, 0, while others may have a second card, 1. Subdevices are on those cards, and HDMI audio is output is often the first, or 0, subdevice. Pulseaudio is configured to use the device "sysdefault" and a chain of processing plugins. If there's an issue with the driver then audio won't run properly on device sysdefault, but there are work arounds. You could edit (as root, sudo gedit) /etc/pulse/default.pa and change the device in line 47 from "device=sysdefault" to "device=hw:0,0" or pethaps "device=hw:0,1" if HDMI is on "hw:0,0". The snd-hda-intel driver should, but often does not, properly configure sound when the computer is made by one if the previously mentioned manufacturers. Here is an extensive list of model options for the snd-hda-intel driver: http://lxr.linux.no/linux+v3.2.19/Documentation/sound/alsa/HD-Audio-Models.txt Remove the default driver and insert the driver with a specific model option. For Dell laptops, try this: sudo su modprobe -r snd-hda-intel modprobe snd-hda-intel model=dell-m6 alsa force-reload For many HP machines this works: sudo su modprobe -r snd-hda-intel modprobe snd-hda-intel model=auto alsa force-reload Then bring up the mixer: alsamixer In alsamixer, use F6 to select your sound card then set levels. The "m" key mutes / unmutes channels. "Esc" to exit alsamixer. Then save the mixer settings, switch to user skywave, and start pavucontrol: alsactl store su skywave pavucontrol In pavucontrol, check the levels again, making sure the speakers or headphone output is unmuted. You should hear sounds doing a speaker test. speaker-test -t sine You should hear a tone alternating between the left and right channels.
0 notes
Text
My First Idris Proof
Tools vs Conveniences
Most of what I do are affected by the tools I use. In writing math my two options are paper and LaTeX. I am a total slob when it comes to paper: loose paper are all over the place and my notebooks are like a homeless graphomaniac's ramblings. That leaves typing in LaTeX, and when it comes to editors I am an editor-agnostic: I'll do emacs, vi, nano, gedit or even ed. If you observe me long enough, you can even see me doing straight cat into a file.
When it comes to programming, I am IDE-averse. Apart from emacs, the last IDE I used was probably Turbo C back in 90's. Because, if my IDE's raison d'etre is to do the boring stuff for me, may be I am using a wrong language to start with. You don't want me to pull a Kant on you:
If I have a book that thinks for me, a pastor who acts as my conscience, a physician who prescribes my diet, and so on--then I have no need to exert myself.
True tools are languages. Editors and frameworks are conveniences.
Diversity, Objectives and Constraints
George Perec once wrote a whole novel on a type-writer whose e-key was removed which was in part inspired by another novel written by an obscure American writer Ernest Wright. In the same vein, if I'd like to write programs that severely restricts my use of side effects then I'd use haskell. If I'd want to do this on the JVM platform then I'd switch to frege, probably. Different objectives and constraints require different tools, and in turn, these constraints might bring out novel ideas you wouldn't normally think of.
In short, learn as much as you can and as diversely as you can. Make choices and write, or cat if appropriate.
Idris Programming Language and Proof Assistants
Idris has been on my todo list for a long time. A strongly and dependently typed functional programming language similar to haskell. I am mainly interested in idris' interactive theorem proving capabilities.
I know that coq is the de facto standard and it does a more comprehensive job than idris, but I like its haskell-like syntax. If you are interested, check Homotopy Type Theory, a machine assisted proof of Feit-Thompson Odd Order Theorem in coq, and a coq proof of the Four Color Theorem.
There are many old, and new and experimental formal proof assistants out there. Here are a few in case you’d want to check out:
Agda
Isabelle
ACL2
Lean
My first proof in Idris
Every journey starts with few small steps. So, here are mine: I am going to verify that the addition in the set of natural numbers is commutative. This proof is different than the commutativity proof given in the official documentation of idris.
The set of natural numbers is given as a recursive type which as a bottom element Z and recursively defined as
data Nat = Z | S Nat
Unlike haskell, the type definitions are preceeded by : instead of :: as you can see below:
unit: (a: Nat) -> plus a Z = a unit Z = Refl unit (S k) = rewrite unit k in Refl
My first lemma is the unitality of the addition. The proof is a simple recursion/induction argument. I think of Refl as the bottom type in proofs which is x=x reflexivity: if you can reduce your proof to that you are done.
The next lemma is the about the fact that (n+1)+m=n+(m+1).
unloop: (a: Nat) -> (b: Nat) -> plus a (S b) = S (plus a b) unloop Z b = Refl unloop (S k) b = rewrite unloop k b in Refl
Then the rest is recursion on the second argument:
comm: (a: Nat) -> (b: Nat) -> plus a b = plus b a comm Z b = rewrite unit b in Refl comm (S k) b = rewrite comm k b in rewrite unloop b k in Refl
0 notes
Link
VI(M) is hard, and sometimes we need to take drastic measures. We understand your needs. Maybe you're new on the job, and you forgot to set your default text editor to nano, emacs, gedit, whatever. VIM pops up and now you have to make a choice...
0 notes