#mean value theorems; Riemann integration
Explore tagged Tumblr posts
Text
Effortpost registry
Kernels and Injectivity; 19 jun 2025
The Topology Game; 2 apr 2025
Graphs as presheaves 4: coverages; 13 feb 2025
The general linear group as a Hopf algebra; 31 oct 2024
Zariski topologies; 14 oct 2024
On integer multiplication and endomorphism algebras; 2 sep 2024
Recommendations for learning category theory; 28 mar 2024
The hairy ball theorem and stably free modules; 11 feb 2024
Topological connectedness and generalized paths; 24 nov 2023
Graphs as presheaves 3: subobject classifiers; 19 oct 2023
Effortpost registry; 18 oct 2023
Graphs as presheaves 2: limits and colimits; 11 oct 2023
Hydrogen bomb vs. coughing baby: graphs and the Yoneda embedding; 7 oct 2023
Extending the D ⊣ U ⊣ I adjunction sequence; 23 sep 2023
The Riemann rearrangement theorem and net convergence; 18 sep 2023
Thoughts on the axiom of choice; 18 feb 2023
Topological spaces and simple graphs as neighbourhood spaces; 15 feb 2023
What is a space?; 10 jan 2023
The exponential function applied to sets; 24 dec 2022
On nilpotent eigenvalues; 23 dec 2022
But IS the empty space connected?; 11 nov 2022
Monads monads monads; 8 nov 2022
Calculating what the triangle identities mean for a bunch of adjunctions and being amazed when it works every time; 7 nov 2022
Defining the Lebesgue integral as a net limit; 27 jul 2022
Rambles about describable sets; 28 oct 2021
Functions with cycling derivatives; 30 aug 2021
Why the rationals have zero length; 31 may 2021
An infinite cardinal valued random variable; 30 may 2021
A field-based functor; 20 mar 2021
Generalized sides; 13 mar 2021
Rambles about metric convexity; 22 feb 2021
Wiggle function convergence; 28 jan 2021
Rambles about infinity; 5 sep 2020
Generalized golf; 24 jun 2020
Rambles about continuousifying series; 10 may 2020
Rambles about being closed under exponentiation; 7 may 2020
Rambles about the groups that come with fields; 3 may 2020
A compilation of donutified functions; 17 mar 2020
Rambles about arithmetic functions; 24 jan 2020
Graphing real functions on a torus >:); 29 nov 2019
78 notes
·
View notes
Note
please help… could you explain integrals to me like i’m five? i barely passed math class by the skin of my teeth in high school and i would really like to get smarter 🙏
ohohoho you've activated my trap card (my ass loves calculus). strap in because this will get a little complex, as is the nature of most calculus.
integrals are conceptually pretty simple in that they do kind of the opposite of derivatives - practically, that means adding up a bunch of stuff to find a result over a stretch of space rather than closing in on an instantaneous point.
the basic idea there is based on an idea called a riemann sum.
what the riemann sum does is estimate the area under a curve by drawing a bunch of rectangles at certain points along the curve, calculate the area of those rectangles, and then add them all up.
the sum assumes each rectangle has the same width, denoted by Δx - this is the total width of the stretch divided by the number of rectangles. the height of each rectangle is given by the value of the function at each of those certain points. Add them up, and we get an approximate of the total area under the curve.

that's a big, long, and kind of annoying way to write it out, so we can rewrite it using sigma notation - basically a way to describe adding a bunch of things up depending on a certain count.
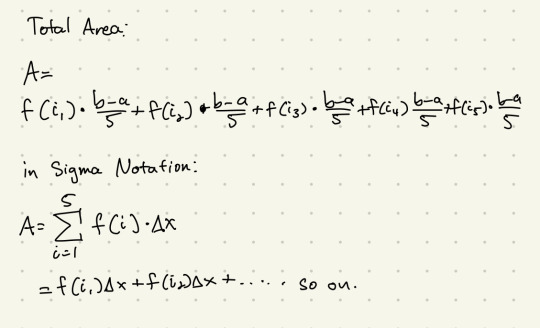
we can make this a general notation for any n number of intervals, like this:

however, the issue remains that no matter how many rectangles we choose to have, we still have some margin of error. so how do we eliminate that error and get the closest possible value for the area under whatever curve we have?
we do what most of calculus is based on: we invite the concept of infinity to take a little tango! what an integral does is it takes the riemann sum and asks it to add up an infinite amount of rectangles. since each rectangle gets infinitely narrower, the margin of error gets infinitely smaller, to the point that we can call the margin of error zero - which is to say, we have a perfectly accurate answer. so that's where we get the limit definition of the integral:

we use dx in the integral to represent Δx, except since the width is infinitely small, we use dx similarly to the notation of derivatives, i.e. an infinitesimally small amount.
as it turns out, the operation of taking an integral is the inverse of taking a derivative - that is to say, they cancel out. this discovery is what nets us an incredibly powerful little tool called the Fundamental Theorem of Calculus

so that's what integrals are and effectively what they do. when it comes to actually evaluating integrals that's a whole different beast, but the basic idea is that the result of an integral is also called an antiderivative, so we reverse the rules we use for derivatives to actually evaluate an integral.
#asks#felix rambles#mathemagics#calculus is my favorite field of math if you couldn't tell#i love it so so dearly#and i jump at any opportunity to explain it to other people
2 notes
·
View notes
Text
IIT JAM Syllabus 2025: A Comprehensive Guide
The IIT JAM (Joint Admission Test for Masters) is one of the most competitive exams for students aspiring to pursue postgraduate studies in esteemed institutions like IITs and IISc. Mathematics, being a core subject, attracts candidates with strong analytical and problem-solving skills. To excel in this exam, a thorough understanding of the IIT JAM Mathematics Syllabus 2025 is essential. This blog outlines the syllabus in detail and provides tips to help candidates prepare effectively.
Overview of IIT JAM Mathematics Syllabus 2025
The IIT JAM Mathematics Syllabus 2025 is crafted to test the candidates' knowledge of fundamental mathematical concepts covered at the undergraduate level. The syllabus is broad, covering topics such as calculus, linear algebra, differential equations, and numerical analysis. Each section focuses on key areas that are crucial for advanced studies and professional applications.
Key Topics in the Syllabus
1. Sequences and Series
This section includes the convergence of sequences and series, tests for convergence (such as comparison, ratio, and root tests), and the study of power series and their radius of convergence.
2. Differential Calculus
Candidates must understand single-variable calculus concepts like limits, continuity, and differentiability. Topics also include Taylor series, mean value theorem, and indeterminate forms. For multivariable calculus, partial derivatives, maxima, minima, saddle points, and the method of Lagrange multipliers are essential.
3. Integral Calculus
This section covers definite and indefinite integrals, improper integrals, and special functions like beta and gamma functions. The application of double and triple integrals is also emphasized.
4. Linear Algebra
A critical area of the syllabus, it focuses on vector spaces, subspaces, linear transformations, rank, nullity, eigenvalues, eigenvectors, and matrix diagonalization. Understanding the solution of systems of linear equations is vital.
5. Real Analysis
This section involves the properties of real numbers, limits, continuity, differentiability, and Riemann integration. Candidates must also be familiar with sequences, Cauchy sequences, and uniform continuity.
6. Ordinary Differential Equations (ODEs)
This includes first-order ODEs, linear differential equations with constant coefficients, systems of linear ODEs, and Laplace transform techniques for solutions.
7. Vector Calculus
Important topics include gradient, divergence, curl, line integrals, surface integrals, and volume integrals, along with Green’s, Stokes’, and Gauss divergence theorems.
8. Group Theory
The basics of groups, subgroups, cyclic groups, Lagrange’s theorem, permutation groups, and homomorphisms are covered.
9. Numerical Analysis
This section focuses on numerical solutions for non-linear equations, numerical integration and differentiation, interpolation methods, and error analysis.
Tips for Preparing the Syllabus
Understand the Weightage: Review past papers to prioritize high-scoring topics like Linear Algebra, Real Analysis, and Differential Calculus.
Strategize Your Study Plan: Divide the syllabus into manageable sections, set achievable goals, and stick to a consistent schedule.
Practice Regularly: Solve previous years’ papers and mock tests to familiarize yourself with the question patterns and improve speed.
Strengthen Fundamentals: Focus on core concepts by revisiting undergraduate textbooks and seeking clarity on challenging topics.
Leverage Online Resources: Utilize tutorials, study materials, and practice tests available online to supplement your preparation.
Conclusion
The IIT JAM Mathematics Syllabus 2025 is extensive yet well-structured, providing a clear framework for aspirants to plan their preparation. By mastering the syllabus and practicing diligently, candidates can confidently tackle the exam and achieve their dream of joining top postgraduate programs. Dedicate time, stay consistent, and focus on strengthening your mathematical foundations to excel in IIT JAM Mathematics 2025.
0 notes
Text
B.Sc Tuition In Noida For Calculus
B.Sc Tuition In Noida For Calculus
B.Sc Tuition In Noida For Calculus
Call CFA Academy For B.Sc Maths Tuition Classes In Noida For Finite, countable and uncountable sets, Real number system as a complete ordered field, Archimedean property; Sequences and series, convergence; Limits, continuity, uniform continuity, differentiability, mean value theorems; Riemann integration, Improper integrals; Functions of two or three variables,…
View On WordPress
#and Gauss divergence theorem.#Archimedean property; Sequences and series#B.Sc Tuition In Noida For Calculus#B.Sc Tuition In Noida For Calculus Call CFA Academy For B.Sc Maths Tuition Classes In Noida For Finite#continuity#convergence; Limits#countable and uncountable sets#differentiability#Directional derivatives#Green&039;s theorem#Improper integrals; Functions of two or three variables#Maxima and minima#mean value theorems; Riemann integration#method of Lagrange&039;s multipliers; Double and Triple integrals and their applications; Line integrals and Surface integrals#Partial Derivatives#Real number system as a complete ordered field#saddle point#Stokes&039; theorem#total derivative#uniform continuity
0 notes
Text
Week 43 to do list
Doing math exercises
Chemistry thermodynamics: read chapter in book
Write down thermodynamics equations
Do thermodynamics exercises
Biochemistry: read chapter about proteins and take notes
MATLAB introduction
Math concept go through:
Riemann sums
Properties of definite integrals
Fundamental theorem of calculus
Mean value theorem for integrals
Intermediate value theorem
Variable substitution
Partial integration
0 notes
Text
10.4 Usubstitution Trig Functionsap Calculus

10.4 U-substitution Trig Functionsap Calculus Answers
10.4 U-substitution Trig Functionsap Calculus Pdf
10.4 U-substitution Trig Functionsap Calculus Problems
10.4 U-substitution Trig Functionsap Calculus Worksheet
Calculus II, Section 7.4, #67 Integration of Rational Functions by Partial Fractions One method of slowing the growth of an insect population without the use of pesticides is to introduce into the population a number of sterile males that mate with fertile females but produce no o spring. Let P represeent. AP Calculus AB Mu Alpha Theta Welcome to AP Calculus AB! Contact me here. Need more review? Browse the Algebra II and Pre-Calculus Tabs. AP ® Calculus AB and BC. COURSE AND EXAM DESCRIPTION. AP COURSE AND EXAM DESCRIPTIONS ARE UPDATED PERIODICALLY. Please visit AP Central. Mathematics 104—Calculus, Part I (4h, 1 CU) Course Description: Brief review of High School Calculus, methods and applications of integration, infinite series, Taylor's theorem, first order ordinary differential equations. Use of symbolic manipulation and graphics software in Calculus. Note: This course uses Maple®.
Math 104: Calculus I – Notes
Section 004 - Spring 2014
10.4 U-substitution Trig Functionsap Calculus Answers
Syllabus
Concept Videos

Skeleton NotesComplete Notes Title More Remainder 10.6, 10.9 Remainder 10.6/10.9 Series Estimation & Remainder Sections 10.8-10.10 Sections 10.8-10.10 Taylor (and Maclaurin) Series Section 10.7 Section 10.7 Power Series Introduction Section 10.6 Section 10.6 Alt. Series Test and Abs. Conv. Conv. Tests Section 10.5 Section 10.5 The Ratio and Root Tests Section 10.4 Section 10.4 The Comparison Tests Section 10.3 Section 10.3 The Integral Test Section 10.2 Section 10.2 Introduction to Series Section 10.1 Section 10.1 Sequences Section 9.2 Section 9.2 Linear Differential Equations Section 7.2 Pt 1Pt 2 Section 7.2 Separable Differential Equations Section 8.8 Section 8.8 Probability and Calculus Odd Ans. Section 8.7 Pt. 1Pt. 2Section 8.7 Improper Integrals L'Hopital Section 8.4 Pt. 1Pt. 2Section 8.4 Partial Fraction Decomposition Section 8.3 Pt. 1Pt. 2Section 8.3 Trig. Substitution Section 8.2 Pt. 1Pt. 2Section 8.2 Integrating Trig. Powers Section 8.1 Pt. 1Pt. 2 Section 8.1 Integration By Parts Section 6.6 Section 6.6 Center of Mass Section 6.4 Section 6.4 Surface Area of Revolution Section 6.3 Section 6.3 Arc Length Section 6.2Section 6.2 Volumes Using Cylindrical Shells Section 6.1 Section 6.1 Volumes Using Cross-Sections disk/washer Review Calc I Review Calc I ReviewLimit, Derivative, and Integral Area b/w CurvesArea b/w Curves Video Example U-substitution Graphs you should know
Print out the skeleton notes before class and bring them to class so that you don't have to write down https://foxspain82.tumblr.com/post/657282647494672384/achievement-unlocked-2watermelon-gaming. Hide paragraph marks in microsoft word for mac. everything said in class. If you miss anything, the complete notes will be posted after class.
10.4 U-substitution Trig Functionsap Calculus Pdf
My Penn Page | Penn Math 104 Page| Penn Undergraduate Math | Advice | Help|
10.4 U-substitution Trig Functionsap Calculus Problems
10.4 U-substitution Trig Functionsap Calculus Worksheet
Version #1 The course below follows CollegeBoard's Course and Exam Description. Lessons will begin to appear starting summer 2020. BC Topics are listed, but there will be no lessons available for SY 2020-2021
Unit 0 - Calc Prerequisites (Summer Work) 0.1 Summer Packet
Unit 1 - Limits and Continuity 1.1 Can Change Occur at an Instant? 1.2 Defining Limits and Using Limit Notation 1.3 Estimating Limit Values from Graphs 1.4 Estimating Limit Values from Tables 1.5 Determining Limits Using Algebraic Properties (1.5 includes piecewise functions involving limits) 1.6 Determining Limits Using Algebraic Manipulation 1.7 Selecting Procedures for Determining Limits (1.7 includes rationalization, complex fractions, and absolute value) 1.8 Determining Limits Using the Squeeze Theorem 1.9 Connecting Multiple Representations of Limits Mid-Unit Review - Unit 1 1.10 Exploring Types of Discontinuities 1.11 Defining Continuity at a Point 1.12 Confirming Continuity Over an Interval 1.13 Removing Discontinuities 1.14 Infinite Limits and Vertical Asymptotes 1.15 Limits at Infinity and Horizontal Asymptotes 1.16 Intermediate Value Theorem (IVT) Review - Unit 1
Unit 2 - Differentiation: Definition and Fundamental Properties 2.1 Defining Average and Instantaneous Rate of Change at a Point 2.2 Defining the Derivative of a Function and Using Derivative Notation (2.2 includes equation of the tangent line) 2.3 Estimating Derivatives of a Function at a Point 2.4 Connecting Differentiability and Continuity 2.5 Applying the Power Rule 2.6 Derivative Rules: Constant, Sum, Difference, and Constant Multiple (2.6 includes horizontal tangent lines, equation of the normal line, and differentiability of piecewise) 2.7 Derivatives of cos(x), sin(x), e^x, and ln(x) 2.8 The Product Rule 2.9 The Quotient Rule 2.10 Derivatives of tan(x), cot(x), sec(x), and csc(x) Review - Unit 2
Unit 3 - Differentiation: Composite, Implicit, and Inverse Functions 3.1 The Chain Rule 3.2 Implicit Differentiation 3.3 Differentiating Inverse Functions 3.4 Differentiating Inverse Trigonometric Functions 3.5 Selecting Procedures for Calculating Derivatives 3.6 Calculating Higher-Order Derivatives Review - Unit 3
Unit 4 - Contextual Applications of Differentiation 4.1 Interpreting the Meaning of the Derivative in Context 4.2 Straight-Line Motion: Connecting Position, Velocity, and Acceleration 4.3 Rates of Change in Applied Contexts Other Than Motion 4.4 Introduction to Related Rates 4.5 Solving Related Rates Problems 4.6 Approximating Values of a Function Using Local Linearity and Linearization 4.7 Using L'Hopital's Rule for Determining Limits of Indeterminate Forms Review - Unit 4
Unit 5 - Analytical Applications of Differentiation 5.1 Using the Mean Value Theorem 5.2 Extreme Value Theorem, Global Versus Local Extrema, and Critical Points 5.3 Determining Intervals on Which a Function is Increasing or Decreasing 5.4 Using the First Derivative Test to Determine Relative Local Extrema 5.5 Using the Candidates Test to Determine Absolute (Global) Extrema 5.6 Determining Concavity of Functions over Their Domains 5.7 Using the Second Derivative Test to Determine Extrema Mid-Unit Review - Unit 5 5.8 Sketching Graphs of Functions and Their Derivatives 5.9 Connecting a Function, Its First Derivative, and Its Second Derivative (5.9 includes a revisit of particle motion and determining if a particle is speeding up/down.) 5.10 Introduction to Optimization Problems 5.11 Solving Optimization Problems 5.12 Exploring Behaviors of Implicit Relations Review - Unit 5
Unit 6 - Integration and Accumulation of Change 6.1 Exploring Accumulation of Change 6.2 Approximating Areas with Riemann Sums 6.3 Riemann Sums, Summation Notation, and Definite Integral Notation 6.4 The Fundamental Theorem of Calculus and Accumulation Functions 6.5 Interpreting the Behavior of Accumulation Functions Involving Area Mid-Unit Review - Unit 6 6.6 Applying Properties of Definite Integrals 6.7 The Fundamental Theorem of Calculus and Definite Integrals 6.8 Finding Antiderivatives and Indefinite Integrals: Basic Rules and Notation 6.9 Integrating Using Substitution 6.10 Integrating Functions Using Long Division and Completing the Square 6.11 Integrating Using Integration by Parts (BC topic) 6.12 Integrating Using Linear Partial Fractions (BC topic) 6.13 Evaluating Improper Integrals (BC topic) 6.14 Selecting Techniques for Antidifferentiation Review - Unit 6
Unit 7 - Differential Equations 7.1 Modeling Situations with Differential Equations 7.2 Verifying Solutions for Differential Equations 7.3 Sketching Slope Fields 7.4 Reasoning Using Slope Fields 7.5 Euler's Method (BC topic) 7.6 General Solutions Using Separation of Variables 7.7 Particular Solutions using Initial Conditions and Separation of Variables 7.8 Exponential Models with Differential Equations 7.9 Logistic Models with Differential Equations (BC topic) Review - Unit 7
Unit 8 - Applications of Integration 8.1 Average Value of a Function on an Interval 8.2 Position, Velocity, and Acceleration Using Integrals 8.3 Using Accumulation Functions and Definite Integrals in Applied Contexts 8.4 Area Between Curves (with respect to x) 8.5 Area Between Curves (with respect to y) 8.6 Area Between Curves - More than Two Intersections Mid-Unit Review - Unit 8 8.7 Cross Sections: Squares and Rectangles 8.8 Cross Sections: Triangles and Semicircles 8.9 Disc Method: Revolving Around the x- or y- Axis 8.10 Disc Method: Revolving Around Other Axes 8.11 Washer Method: Revolving Around the x- or y- Axis 8.12 Washer Method: Revolving Around Other Axes 8.13 The Arc Length of a Smooth, Planar Curve and Distance Traveled (BC topic) Review - Unit 8
Unit 9 - Parametric Equations, Polar Coordinates, and Vector-Valued Functions (BC topics) 9.1 Defining and Differentiating Parametric Equations 9.2 Second Derivatives of Parametric Equations 9.3 Arc Lengths of Curves (Parametric Equations) 9.4 Defining and Differentiating Vector-Valued Functions 9.5 Integrating Vector-Valued Functions 9.6 Solving Motion Problems Using Parametric and Vector-Valued Functions 9.7 Defining Polar Coordinates and Differentiating in Polar Form 9.8 Find the Area of a Polar Region or the Area Bounded by a Single Polar Curve 9.9 Finding the Area of the Region Bounded by Two Polar Curves Review - Unit 9
Unit 10 - Infinite Sequences and Series (BC topics) 10.1 Defining Convergent and Divergent Infinite Series 10.2 Working with Geometric Series 10.3 The nth Term Test for Divergence 10.4 Integral Test for Convergence 10.5 Harmonic Series and p-Series 10.6 Comparison Tests for Convergence 10.7 Alternating Series Test for Convergence 10.8 Ratio Test for Convergence 10.9 Determining Absolute or Conditional Convergence 10.10 Alternating Series Error Bound 10.11 Finding Taylor Polynomial Approximations of Functions 10.12 Lagrange Error Bound 10.13 Radius and Interval of Convergence of Power Series 10.14 Finding Taylor Maclaurin Series for a Function 10.15 Representing Functions as a Power Series Review - Unit 8
Version #2 The course below covers all topics for the AP Calculus AB exam, but was built for a 90-minute class that meets every other day. Lessons and packets are longer because they cover more material.
Unit 0 - Calc Prerequisites (Summer Work) 0.1 Things to Know for Calc 0.2 Summer Packet 0.3 Calculator Skillz
Unit 1 - Limits 1.1 Limits Graphically 1.2 Limits Analytically 1.3 Asymptotes 1.4 Continuity Review - Unit 1
Unit 2 - The Derivative 2.1 Average Rate of Change 2.2 Definition of the Derivative 2.3 Differentiability (Calculator Required) Review - Unit 2
Unit 3 - Basic Differentiation 3.1 Power Rule 3.2 Product and Quotient Rules 3.3 Velocity and other Rates of Change 3.4 Chain Rule 3.5 Trig Derivatives Review - Unit 3
Unit 4 - More Deriviatvies 4.1 Derivatives of Exp. and Logs 4.2 Inverse Trig Derivatives 4.3 L'Hopital's Rule Review - Unit 4
Unit 5 - Curve Sketching 5.1 Extrema on an Interval 5.2 First Derivative Test 5.3 Second Derivative Test Review - Unit 5
Unit 6 - Implicit Differentiation 6.1 Implicit Differentiation 6.2 Related Rates 6.3 Optimization Review - Unit 6
Unit 7 - Approximation Methods 7.1 Rectangular Approximation Method 7.2 Trapezoidal Approximation Method Review - Unit 7
Unit 8 - Integration 8.1 Definite Integral 8.2 Fundamental Theorem of Calculus (part 1) 8.3 Antiderivatives (and specific solutions) Review - Unit 8
Unit 9 - The 2nd Fundamental Theorem of Calculus 9.1 The 2nd FTC 9.2 Trig Integrals 9.3 Average Value (of a function) 9.4 Net Change Review - Unit 9
Unit 10 - More Integrals 10.1 Slope Fields 10.2 u-Substitution (indefinite integrals) 10.3 u-Substitution (definite integrals) 10.4 Separation of Variables Review - Unit 10
Unit 11 - Area and Volume 11.1 Area Between Two Curves 11.2 Volume - Disc Method 11.3 Volume - Washer Method 11.4 Perpendicular Cross Sections Review - Unit 11

0 notes
Text
Ap Calculushome

Course Outline
Ap Calculus Homework Help
Ap Calculus Parametric Functions Homework
This course covers all of the topics required for the AP Calculus AB exam.
Welcome to AP Calculus (AB) for the 2012-13 school year! Here is where you'll find all the necessary information for upcoming material being covered, as well as material and assignments previously covered. Ap calculus ab Calculators I recommend: TI 84 Plus Silver Edition,TI 84 Plus Silver Edition Color, TI 89. (Note: The TI 89 and TI nspire-CAS are not approved for the IB or ACT Test). Mcculloch mac 3200 chainsaw manual pdf. The AP Calculus Exam is Tuesday May 15,2018 at 7:30 am! BE PREPARED!!!!! Please note the following websites! Calculus AB Bible Paul's Online Notes Khan Academy videos Pre-Cal Review AP Exam Prep Ideas.
Ap Calculus Homework Help
Chapter 1: Introduction The problems that Calculus solves, introduction to derivatives, finding rates of change from graphs, from equations, and from data, Numerical derivatives, Introduction to Integrals, Approximating integrals from graphs, from equations and from data, the Trapezoid Rule
Chapter 2: Limits A graphical approach to limits, Describing function behavior with limits, Asymptotes, Rational Functions, Polynomial end behavior, The Limit Theorems, Evaluating limits, Continuity, The Intermediate Value Theorem
Chapter 3: Derivatives A graphical look at derivatives, Difference Quotients, the Derived Function, Notation, Numerical calculations of derivatives, Tangents and Linear Approximation, Differentiability and Continuity, the Chain Rule, the Product Rule, the Quotient Rule, Leibniz' Proofs, Derivatives of Trig Functions, Implicit Differentation, Derivatives of Inverse Functions, Derivatives of Inverse Trig Functions
Chapter 4: Applications of Derivatives The Extreme Value Theorem, Rolle's Theorem and the Mean Value Theorem, First and Second Derivatives, Concavity and Inflection Points, Graphs and Curve Sketching, The Calculus of Motion, Max-Min problems, Related Rates, Practice
Chapter 5: Integrals Antiderivatives, Integrals, Infinitesimals, Riemann Sums, Definite Integrals, The Fundamental Theorem of Calculus, Properties of Definite Integrals, Numerical Methods, Integration by Substitution, Average Value
Chapter 6: Exponential Functions and Differential Equations Derivatives of exponential functions, Derivatives of logarithmic functions, Derivatives and integrals of base b exponents, Integrals with variable limits, Logarithmic Differentiation, Integrals of trig functions, Intro to Differential Equations, Examples and applications, Slope Fields, Euler's Identity
Chapter 7: Applications of Integrals The area of a plane region, The Calculus of Motion, Real world applications, Integrating to find volumes, Plane Slicing, Solids of Revolution, Cylindrical Shells
Course Outline This course covers all of the topics required for the AP Calculus AB exam.

Ap Calculus Parametric Functions Homework
Chapter 1: Introduction The problems that Calculus solves, introduction to derivatives, finding rates of change from graphs, from equations, and from data, Numerical derivatives, Introduction to Integrals, Approximating integrals from graphs, from equations and from data, the Trapezoid Rule
Chapter 2: Limits A graphical approach to limits, Describing function behavior with limits, Asymptotes, Rational Functions, Polynomial end behavior, The Limit Theorems, Evaluating limits, Continuity, The Intermediate Value Theorem
Chapter 3: Derivatives A graphical look at derivatives, Difference Quotients, the Derived Function, Notation, Numerical calculations of derivatives, Tangents and Linear Approximation, Differentiability and Continuity, the Chain Rule, the Product Rule, the Quotient Rule, Leibniz' Proofs, Derivatives of Trig Functions, Implicit Differentation, Derivatives of Inverse Functions, Derivatives of Inverse Trig Functions
Chapter 4: Applications of Derivatives The Extreme Value Theorem, Rolle's Theorem and the Mean Value Theorem, First and Second Derivatives, Concavity and Inflection Points, Graphs and Curve Sketching, The Calculus of Motion, Max-Min problems, Related Rates, Practice
Chapter 5: Integrals Antiderivatives, Integrals, Infinitesimals, Riemann Sums, Definite Integrals, The Fundamental Theorem of Calculus, Properties of Definite Integrals, Numerical Methods, Integration by Substitution, Average Value
Chapter 6: Exponential Functions and Differential Equations Derivatives of exponential functions, Derivatives of logarithmic functions, Derivatives and integrals of base b exponents, Integrals with variable limits, Logarithmic Differentiation, Integrals of trig functions, Intro to Differential Equations, Examples and applications, Slope Fields, Euler's Identity
Chapter 7: Applications of Integrals The area of a plane region, The Calculus of Motion, Real world applications, Integrating to find volumes, Plane Slicing, Solids of Revolution, Cylindrical Shells

0 notes
Text
BIOLOGICAL SCIENCES
NOTE: PRACTICALLY STUDENTS AND INSTRUCTORS/PROFESSORS WILL MAKE USE OF MULTIPLE SOFTWARE AND DATA SOURCES OUT OF THE LISTS PROVIDED FOR COURSES AND ACTIVITIES. A PARTICULAR SOFTWARE OR DATA SOURCE HAS ITS RESPECTIVE FOCUSES AND STRENGTHS. Biology degree pursuits concern environments that invite, assimilate, nurture and sustains advancement beyond cultural and racial settings. The biology environment is not a place for de-facto vice sororities nor vice fraternities. The biology environment concerns cerebral functional growth, ingenuity, adaptation and advancement in laboratory activity and technology from acquired knowledge and skills. BIOLOGY REALM Courses GENERAL BIOLOGY I & II, GENERAL CHEMISTRY I & II, ORGANIC CHEMISTRY I & II, to be accompanied by lab instruction; other courses with given syllabuses to have lab instruction if expressed. Of interest: https://www.raspberrypi.org/magpi/digital-microscope/
Calculus for Biological Sciences I Limits, continuity, derivatives, mean value theorem, extrema, curve sketching, related rates, differentiation of the trig, log, and exponential functions, basic integration techniques, with particular motivations from and application to the Biological Sciences. Mandatory enrolment for beginner freshmen Typical Text: Calculus for Biology and Medicine, by Claudia Neuhauser, Pearson Course to be taught with use for scientific/graphing calculators and immersion into RStudio Problem Sets --> Problem Sets: There will be (mostly) weekly homework assignments. Exams --> There will be 2 midterms at the end of week 4, and at the end of week 8. There will be a two-hour final exam at the time scheduled by the registrar’s final exam calendar. Topics List: 1.2 Elementary Functions 1.3/2.1 Graphing/Exponential Growth and Decay 2.2 Sequences 3.1-3.4 Limits and Continuity 3.5 Properties of Continuous Functions 4.1 Derivatives 4.2-4.3 Rules of Differentiation, Product and Quotient Rules 4.4 Chain Rule and Higher Derivatives 4.5-4.7 Derivatives of Special Functions and Inverse Functions 5.1-5.3 Extrema, Mean Value Theorem, Monotonicity, Concavity, Inflection Points 5.4 Optimization 5.5 L'Hospital's Rule 5.8 Antiderivatives 6.1 The Definite Integral 6.2 The Fundamental Theorem of Calculus 6.3 Applications of Integration 7.1-7.2 Integration Techniques Also: Small-group Projects Calculus for Biological Sciences II Mandatory enrolment for upper level freshmen granted that they have successful completed Calculus for Biological Sciences I Learning Outcomes: in this class we will learn how to -Find the family of antiderivatives (if possible) for a continuous function -Approximate definite integrals using Riemann sums -Use substitution and integration by parts to compute indefinite and definite integrals -Compute and interpret definite integrals with finite and infinite limits of integration -Set up single and coupled differential equations based on written descriptions including predator/prey models, population ecology, competitive selection, and chemical exchange across a membrane -Solve certain pure-time, autonomous, and non-autonomous differential equations using integration and separation of variables -Find and determine the stability of equilibria in autonomous differential equations; draw relevant phaseline diagrams -Sketch solutions to single and coupled differential equations from an initial condition -Verify solutions to, and use Euler’s method with, differential equations in two dependent variables -Use nullclines and find equilibria of systems of differential equations in two dependent variables -Sketch phase-plane trajectories for systems of differential equations Typical Text: Text: Calculus for the Life Sciences, by Frederick Adler. Course to be taught with use for scientific/graphing calculators and immersion into RStudio Problem Sets --> Will be (mostly) weekly homework assignments. Quizzes --> We have 4-5 quizzes Exams --> There will be 2 midterms at the end of week 4, and at the end of week 8. There will be a two-hour final exam at the time scheduled by the registrar’s final exam calendar. Approximate Schedule --> Week 1: Differential equations and antiderivatives. (4.1-4.2.) Week 2: Integration: how to compute antiderivatives. (4.3-4.4.) Week 3: Definite integrals. Applications of integrals. (4.5-4.6.) Week 4: Improper integrals. (4.7.). Midterm I. Week 5: More complicated (and interesting) differential equations. 5.1-5.2. Week 6: Autonomous differential equations. (5.3-5.4.) Week 7: Differential equations in dimension 2. (5.5-5.6.) Week 8: Solving DEs graphically or approximately. (5.7.). Midterm II. Week 9: The dynamics of a neuron. (5.8.) Week 10: Review. Week 11: Final exam A. MICROBIOLOGY degree. Apply additional lab instruction or compliment lab hours wherever needed for courses. Students will proceed with courses based on prerequisites they have successfully completed with satisfactory grade requirement. Curriculum: --Core Courses Scientific Writing I & II, General Biology I & II, General Chemistry I & II, Organic Chemistry I (with labs), Biochemistry (with labs), Organic Synthesis Laboratory, Biostatistics I & II, Advanced Statistical Modelling and Machine Learning for Biostatistics --Professional Necessities Cell Biology (with labs), Molecular Biology I & II, Microbiology I & II (with labs), Environmental Microbiology, Clinical Microbiology, Microbiology of the Digestive System, Bacteriology, Virology, Tissue Culture & Virology Lab, Genetic Engineering & Technology, Comparative Genomics (check METAB BIO), Biotechnology Laboratory, Advanced Biotechnology Laboratory, Tissue Engineering, Microbiology Research --Mandatory Courses Calculus for the Biological Sciences I & II, ODE, General Physics I Note: if students are interested in Molecular Biology II, such course to be electives, granted that student have the academic time to pursue them. Description of particular listed courses: Biostatistics I Course concerns probability and statistics applied to problems in biology, industrial/occupational health, and epidemiology. Use of statistical software R for data analysis is emphasized extensively. Note: this course is designed for students majoring in the biological sciences with a second term calculus background. Through the extensive use of practical examples, this course is expected to motivate and teach students statistics knowledge that would be helpful for their major study. The computer program R is the standard statistical program for this course. Students will use R to complete data analysis projects. R can be downloaded and installed on your personal computer for free following instructions at http://www.r-project.org/. In addition, the R environment will be augmented by RStudio interface with other R packages. This course covers fundamental concepts in probability and statistics, including data description, design of experiment, probability rules and distributions, statistical inference and linear regression. Definitions will be learned through real-world examples and applications. Besides these traditional materials and subjects, topics and methods that are particularly applicable to the biological sciences will be introduced. Again, much focus on the applications of statistical ideas to realistic data and practices. Students are expected to use materials learned from this course to guide statistical practice for their major studies in the future. After successfully completing this course the expectation is that students will be able to: 1. To grasp concepts in probability and be able to apply basic probability rules, distributions, and laws to solve conceptual statistics questions 2. Use statistical guidelines and common sense to interpret the process of data collection, description and analysis, and to design statistically sound experiments 3. Learn various statistical inference techniques and be able to select appropriate methods for specific data sets and scientific purpose 4. Link the course materials with real-life examples, and explore the opportunities for other biological applications 5. Interpret statistical reports and carrying out data analysis using R. Several data projects will be assigned during the semester. Independent work is expected. This is not a course of “pen and paper finesse” succeeding the composition of gunk and bamboozle on a writing board. One can’t be successful in statistics by only writing down theory. Practice with an environment that applies intelligence and engagement is essential. There’s no point in doing statistics if one doesn’t know how to acquire and manipulate real data. Real data realistically outnumbers the fingers and toes one possesses. Most of grading will be based on projects (having commentary descriptions) accompanied by the analytical process development description done in a word processor. Typical Texts --> Will make use of R language Statistics texts under CRC Press and Springer publishing Tools --> R language and R Studio Note: a calculator at times may prove useful for the idealistic or the “synthetic” customary questions. “R Monograph Notebook” --> Students should maintain a notebook as they proceed through the course and learn how to do analyses in R. This assignment involves a notebook that lists the syntax and provides a brief explanation of each function that students learn during the course. Instructor will assign R maturity development questions to be in tune with course progress; you will only be allowed to use your monograph to assist with assigned questions. The notebook will be handed in near the end of the semester and handed back to the students after grading. Such a notebook can be an extremely useful resource both during and after the course to quickly refresh one’s memory on the details of a particular function. Design with R most likely will vary among students. Poor development in a such a notebook may or may not correlate with poor grades. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS. Grading --> Problem Sets 20% R Labs 25% R Activities 0.7 Will involve all course topics R Monograph Lab Notebook 0.3 R maturity development questions End of term lecturer observation 3 Exams 30% Assigned Projects 25% Exams --> Limited open notes. I don’t like setting up myself and students for embarrassment; you are not perfect with statistics, so expect exams to be primarily knowledge based and the calculus related fodder. Most of your development will come from homework and labs; it is what it is. Note: limited open notes. Students may be more comfortable with certain R packages. Again --> Several data projects will be assigned during the semester. Independent work is expected. Course Outline --> Introduction to statistics, data and R Statistical Measures and Summary Statistics for data sets Data acquisition: Sources/databases (ecological & biological), file types, APIs, etc. Data Wrangling Summary Statistics Applied probability theory Axioms of probability Modelling frequencies and establishing densities Simulating random variables from real experiments Probability distributions and properties Law of Large Numbers and the Central Limit Theorem Introduce the Law of Large Numbers (LLN) Central Limit Theorem (CLT). Identify Exponential, Poisson and Binomial data and respectively determine in a manner to confirm LLN and CLT. Routledge, R., Chebyshev’s Inequality, Encyclopaedia Britannica Is there too much reliance on assuming normal or Gaussian distribution? Towards Chebyshev’s inequality what amount of repetition (regarding LLN and CLT) of an experiment is adequate towards Chebyshev becoming relevant? Overview and goals of various concentration inequalities (just a survey). Sample Estimates Chi-square distribution The bottom line is to establish the flow of the uses competently with applications involving real raw data. Comprehending categorical data sets Organisation of data and sensitivity of categories concerning traits of interest. Test for independence McHugh ML. (2013). The chi-square Test of Independence. Biochem Med (Zagreb). 23(2): 143-9. Test of homogeneity Test of variance Applications of the Chi-Square distribution with confidence intervals T-distribution Kim T.K. & Park J. H. (2019). More About the Basic Assumptions of T-test: Normality and Sample Size. Korean J Anesthesiol. 72(4): 331-335. Sample size determination Population parameter estimation Confidence intervals Directly logistical to understand what you’re doing in R F-distribution Assumptions for the F-distribution Relevance to the biological sciences (active data immersion) Note: not textbook finesse, rather how and when actively. Goodness of fit: fit of distributions Summary Statistics Skew and Kurtosis P-P and Q-Q Statistical Tests Definition, Null hypothesis One-sided & two-sided tests of hypothesis Types of test statistics Comprehending critical values for ideal distributions Significance levels Critical values for real raw data sets Does your data exonerate ideal distributions? Chi-Square Test Kolmogorov-Smirnov Test Anderson-Darling test Shapiro-Wilk Test MLE and Method of Moments Manual tasks will be limited to at most 4 element data sets Computational logistics for large data sets followed by implementation Review/probe data for goodness of fit module for appropriate distribution You may be tasked with distribution determination before parameter/point estimation Hypothesis testing (exploratory, sans assumption of distribution) Note: as aspiring biologists I can’t give “zombie textbook problems” and expect you to relate to a profession tangibly and fluidly. You will be exposed to raw professional data from various sources. You will develop the four mentioned steps. You should ask yourselves if the hypotheses are practical as well. Why is normal distribution assumed? Will be exploratory rather than zombie problems. Namely, knowledge and skills from Goodness of fit module. Then proceed with the following: 1.State the two hypotheses so that only one can be right 2.Formulate an analysis plan, outlining how data will be evaluated 3.Carry out the plan and physically analyze the sample data 4.Analyse the results and either reject the null, or state that the null is plausible, given the data 5.Directly logistical to understand what you’re doing in R Test of Proportions (exploratory) Differences in population (mean and median) Comparisons of variances Confidence limits for means. Does it require normality? Correlation (includes misuse of types and resolutions) Pearson Correlation Crucial Conditions Structure Implementation Spearman Correlation Crucial Conditions Structure Implementation Kendall Correlation Generating heat maps. The ggpairs() function Bivariate Regression Multiple Regression Model components Methods to select variables OLS MUST: Summary Statistics and forecasting Analysis of Covariance Must be exploratory, else it’s toxic Non-parametric statistics Resampling methods Falsified Data Hartgerink C, Wicherts J, van Assen M (2016). The Value of Statistical Tools to Detect Data Fabrication. Research Ideas and Outcomes 2: e8860. Al-Marzouki, S., Evans, S., Marshall, T., & Roberts, I. (2005). Are these data real? Statistical methods for the detection of data fabrication in clinical trials. BMJ, 331(7511), 267–143. Yamamoto, K., & Lennon, M. L. (2018). Understanding and Detecting Data Fabrication in Large-Scale Assessments. Quality Assurance in Education, 26(2), 196–212. Prerequisites: General Biology II, Calculus II Biostatistics II --This succeeding course in the sequence will have more emphasis on incorporating journal articles and real world experiments. --Students will have to orchestrate inquisitions by exploratory data analysis and statistical methods involving R. There will be assigned data sets and journal articles to do just that. --This is not a “pen and paper course”. Texts and journal articles will cater to subjects both from prerequisite and this course. Means of data retrieval and manipulation are crucial; it may be the case that the data desired in inaccessible, hence students will have to resort to alternative data sources that yields much different conclusions. NOTE: personally refresh your knowledge and acquired R skills from calculus and Biostatistics I alongside ordained reacquaintances in course. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS, AND KEEP YOUR DAMN BUSINESS . Assessment --> Assignment Sets (prerequisite & current level from multiple sources) 15% Analytical and R based 3 Exams (prerequisite & current level) 30% Labs + Data Analysis Term Project 40% 2 Field Inquisitions with R 15% Conducted Journal Articles Computational Inquisitions Supporting data sets to be provided Gov’t administration field experiments inquisitions Assignment Sets --> Will be reacquainted with prerequisite tasks, prerequisite projects AND current level tasks (analytical and R computational). Exams --> Exams will have the same manner of administration and activities as exams from prerequisites. Yet, consisting of both prerequisite tasks AND current level tasks. Limited open notes. LABS WITH R --> Data Processing Data Assimilation (ecological & biological), file types, APIs, etc. Data Wrangling Descriptive Statistics Hypothesis Testing (advance practice from prerequisite) Expt. Design, Multiple Comparisons Regression (Mult. Reg. and Dummy) ANCOVA MANCOVA Non-parametric Statistics (advance practice from prerequisite) Clustering PCA and Kernel PCA Comparing & Averaging Models Analysis of Trait Evolution Fitting models of Trait Evolution TERM PROJECT --> The term project has been broken down into multiple components due throughout the semester to provide further guidance for students. On given date, students will select a dataset to use for their term project. Students can either provide their own dataset (if they have collected data during their research), or will be given the opportunity to analyse a complex dataset supplied by faculty as their term project. For the Hypothesis Activity (given due data), students will take a close look at their dataset and formulate biological hypotheses that they would like to test statistically. The assignment will be handing in these hypotheses. For the Experimental Design assignment (given due), students will outline which analyses they will use to test their biological hypotheses and provide the specific explicit statistical hypotheses that they will test. The Term Project Report (given due having additional 1 week collection buffer) will be written after students complete their analyses. The report will include a Statistical Methods and a Results section, complete with tables and figures. Methods should include sufficient detail to redo the analyses. The results should include everything necessary for interpretation of their analyses and data, but not superfluous material. Term Project Reports for all students should include a title page with a title, student name, course number and name, and assignment name. The text of the report should be double spaced, with indented paragraphs, 1” margins, 12pt Times New Roman Font, and page numbers. Tables should be single spaced with headings above each table. Figures should have captions below each figure. Figures and tables can be embedded in the text or provided at the end of the document. Literature cited should follow the format for the journal Evolution. Assignments that do not follow these formatting instructions will be returned to the student for correction prior to grading. NOTE: most labs done serve as structure for your HA and ED NOTE: I will be collecting your R development (having sensible commentary) for the term project in PDF along with the term report in PDF. Finally, students will give a short, in-class presentation about their study, analyses and findings. Presentations will be in PowerPoint. MAJOR COURSE TOPICS --> Methods of data acquisition, data wrangling and summary statistics (prerequisite reinforcement) Goodness of Fit (prerequisite reinforcement) MLE and MoM Hypothesis Testing (prerequisite reinforcement) Experimental Design & Sampling Regression (multivariate) OLS review (variable selection, summary statistics, forecasting) Quantile Regression compared to Least Squares regression Variable selection, summary statistics, forecasting ANCOVA MANCOVA Non-parametric Statistics Resampling Techniques Clustering (K-means or DBSCAN?) Principal Component Analysis (PCA) and Kernel PCA Model Selection & Likelihood (emphasis on computational logistics and implementation) Phylogenetic Regression Extensions of Phylogenetic Statistics Prerequisite: Biostatistics I Advanced Statistical Modelling and Machine Learning for Biostatistics This course explores advanced statistical modeling techniques and machine learning methods as applied to biostatistical problems. Topics include generalized linear models, hierarchical modeling, Bayesian statistics, and the integration of machine learning algorithms for analyzing complex biological and health data. Note: 2 lectures per week, with approximately 2 hours per lecture. Assignments -- Assignments will be quite laborious in the interest of sustainability with knowledge and skills through your journey in biostatistics. Each assignment will comprise of the following elements: A. problems and tasks encountered in both Biostatistics I & II. Such problems and tasks will also make extensive use of R. As well, being advanced biostatistics students, projects from Biostatistics I & II can/will also be considered basic assignments as well. B. Course level assignments to such given course topics. Good emphasis on ability to comprehend and specify the transition from prerequisite skills/tools to course level tools/method/skills; then implementation. Will also make extensive use of R. Data Science Basics Quizzes -- For the Data Science Basics module there will be handwritten quizzes to test knowledge, comprehension, appropriateness and T/F. Exams -- Exams will account for all modules. Assignments will be strong foresight of what’s to appear on exams. You will be making extensive use of R with open notes for all course modules. Exams will feel like projects where each “project” will involve multiple modules. Make-up Student Project -- Applying Advanced Models to Biostatistical Data. Concerns students who are interested in making up lost weight towards their final grade; the better you did in this course, the lower the value. Can regain up to 5% for final grade. Students will be given a sack to randomly (and blindly) pick a project. Students will have until 2 days before the final grade submission deadline to submit projects. Students will be privately given project details via student email where they will have to acquire the data from specified sources. Course Assessment -- Assignments 20% 3 Exams for all modules 60% 3 Data Science Basics Quizzes 15% Will be precursors to exam(s) for the Data Science Basics module Make-up Student Project (being conditional) COURSE OUTLINE -- WEEK 1-3. Introduction to Generalised Linear Models (GLMs) with model estimation and summary statistics Multilinear Regression (fast fast review) Quantile Regression (fast review) Logistic Regression Poisson Regression WEEK 4-5. Hierarchical Modelling (HM) Introduction to HM Multilevel Modelling Random Effects and Mixed Models WEEK 6-7. Bayesian Statistics in Biostatics Note: I don’t introduce things to be a disgusting, miserable, viral bastard. Module will be extremely goal oriented, namely, problem, goal(s), methodology, logistics, implementation, evaluation. No social nor psychological probes/inquisitions upon students; there are certified licensed professionals elsewhere tied to meaningful or economic interests. Bayesian Inference (to the point, constructive and economical) Markov Chain Monte Carlo (MCMC) Methods – only constructive and economical methods Bayesian Regression Models WEEK 8-9. Advanced GLMs and Extensions Negative Binomial Regression Zero-Inflated Models Generalised Estimating Equations (GEE) WEEK 10-15. Data Science Basics Note: subjects of overfitting or underfitting arise in model validation statistics. Data Acquisition. Data Probing: glimpse(), str() Data Wrangling (functions from dplyr R package with piping) Summary Statistics, Skew, Kurtosis, Correlation Analysis and Heatmaps Machine Learning Overview Feature Selection with R (underlying methods may not be fully comprehended, but that’s generally the world): Principal Component Analysis (PCA), Kernel PCA, Boruta, FSelectorRccp. Comparative observation among such prior three also expected. Multiple Regression (very rapid review) OLS and Quantile Classification Logistic Regression Support Vector Machines Decision Trees Random Forests Clustering (K-Means and DBSCAN advanced repetition) Includes the Elboew Method, Silhouette Score and Davies-Bouldin Index Prerequisites: General Biology II, General Chemistry II, Biostatistics II Organic Chemistry I In-depth study of: (i) the structure of organic compounds and the functional groups (bonding, acid-base properties, nomenclature, conformations, stereochemistry), and (ii) the synthesis and reactivity (including detailed mechanisms) of alkanes, alkenes, alkynes, halides, alcohols, ethers, epoxides, sulfides and organometallic reagents. Laboratory experiments are related to topics covered in lecture and emphasize organic laboratory techniques, synthesis and spectroscopic characterization of organic molecules. Typical Texts: McMurry, John E. Organic Chemistry. 8th Edition. Brooks/Cole, 2012. McMurry, Susan. Study Guide with Student Solutions Manual. 8th Edition. Brooks/Cole. Typical Lab Manual: Barbaro, John and Richard K. Hill. Experiments in Organic Chemistry. 3rd Edition, Contemporary Publishing Company of Raleigh, Inc., 2006 Grading: 3 Exams (50% combined) Cumulative Final Exam (25%) Labs (25%) (On the occasion of significant improvement on the final exam, more weight will be placed on the final exam) INSTRUCTIONAL METHODS: List the different instructional methods you might use, in the course of the semester. List supplementary learning options, if any: Traditional lecture with use of chalkboard Computer assisted diagrams and graphics Molecular Models Team work in the laboratory Homework assignments Solving specific questions related to content studied Written exams and distribution of study questions/previous exams Use of the Internet UNIQUE ASPECTS OF COURSE (such as equipment, specified software, space requirements, etc.): Organic chemistry laboratories and their associated equipment, instruments and chemicals. Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Ch. 1 Structure and Bonding Bonding; Hybridization; Drawing Chemical Structures; Functional Groups; Intro to IR Spectroscopy Ch. 2 Polar Covalent Bonds; Acids and Bases Chemical Bonding (Ionic and Covalent); Electronegativity and Dipole Moments; Formal Charges; Resonance Structures; Acid Base Theory (Bronsted-Lowry, Lewis); Acid and Base Strength (pKa); Acid-Base Reactions; Organic Acids and Organic Bases Ch. 3 Organic Compounds: Alkanes and their Stereochemistry Alkanes, Alkane Isomers, and Alkyl Groups; Properties of Alkanes; Conformations Ch. 4 Organic Compounds: Cycloalkanes and their Stereochemistry Cis-Trans Isomerism in Cycloalkanes; Stability and Conformations of Cycloalkanes; Chairs Ch. 5 Stereochemistry at Tetrahedral Centres Enantiomers, the Tetrahedral Carbon and Chirality; Optical Activity; R/S Sequence Rules; Diastereomers and Meso Compounds; Racemic Mixtures, Resolution of Enantiomers; Prochirality; Chirality in Nature Ch. 6 An Overview of Organic Reactions Kinds of Organic Reactions (Radical and Polar); Mechanisms; Describing a Reaction (Equilibria, Rates, Energy Changes, Bond Energy; Transition States, and Intermediates) Ch. 7 Alkenes: Structure and Reactivity Preparation and use of Alkenes; Cis-Trans Isomerism; Alkene Stereochemistry and E/Z Designation; Stability of Alkenes; Electrophilic Addition Reactions; Markovnikov’s Rule: Carbocation Structure and Stability; Carbocation Rearrangements Ch. 8 Alkenes: Reactions and Synthesis Preparation of Alkenes via Elimination Reactions; Addition Reactions of Alkenes (Halogenation, Hydration, Halohydrins, and Hydrogenation); Oxidation of Alkenes (Epoxidation and Hydroxylation); Addition of Carbenes; Radical Additions to Alkenes (Polymer Formation); Reaction Stereochemistry Ch. 9 Alkynes: An Introduction to Organic Synthesis Preparation of Alkynes; Addition Reactions of Alkynes (X2, HX, H2O, H2); Oxidative Cleavage; Alkyne Acidity and Alkylation; Introduction to Organic Synthesis Ch. 11 Reactions of Alkyl Halides: Nucleophilic Substitutions and Eliminations SN2, SN1, E2, E1, E1cB Reactions; Zaitsev’s Rule; Deuterium Isotope Effect Ch. 10 Organohalides Preparation of Alkyl Halides and Grignards; Radical and Allylic Halogenation; Organic Coupling Reactions, Redox in Organic Chemistry Ch. 17 Alcohols and Phenols Properties of Alcohols and Phenols; Preparation and Reactions of Alcohols; Reactions of Phenols Ch. 18 Ethers and Epoxides; Thiols and Sulfides Synthesis and Reactions of Ethers; Cyclic Ethers (Epoxides); Reactions of Epoxides: Crown Ethers; Thiols and Sulfides LABS --> Some experiments require more than one lab period to complete. Based on an instructor’s preference, availability of equipment/supplies or constraints within a given semester, this laboratory schedule is subject to change, including but not limited to, the addition or replacement of one or more of the above experiments with the following experiments: Addition of Bromine to E-Cinnamic Acid in Methylene Chloride Substitution Reactions of Alkyl Halides: Relative Rates Triphenylmethanol with Hydroiodic Acid 1. Check-in, Laboratory Safety, Practices and Waste Disposal. Simple Distillation. 2. Spectroscopy: Introduction to Infrared Spectroscopy. 3. Recrystallization, IR and Melting Point of benzoic acid. 4. Extraction of Organic Compounds from Natural Sources: Trimyristin from Nutmeg. 5. Paper Chromatography 6. Dehydration of Cyclohexanol. 7. Dimerization of 2-Methylpropene 8. Preparation of Diphenylacetylene Starting from Trans-Stilbene. 9. Preparation of Butyl Bromide/Preparation of t-Butyl Chloride (SN2/SN1). 10. Oxidation of Isoborneol to Camphor. 11. The Williamson Ether Synthesis: Preparation of Aryloxyacetic Acid from Cresol. Prerequisites: General Chemistry II Organic Synthesis Laboratory Practice of organic laboratory techniques. Three hours of laboratory per lab session, twice a week. Approved chemical safety goggles meeting whatever national standards. The purpose of this laboratory course is to introduce students to the techniques that organic chemists (as well as biochemists, physical chemists, etc.) use in their daily routines. After learning and understanding those techniques, students will apply their knowledge to new situations to understand synthesis reactions, molecular structure determination, and analysis of (un)known compounds. Organic chemistry laboratory is important for several reasons. It introduces students to many different laboratory practices and concepts that will be used in subsequent chemistry laboratory classes and in other laboratory situations in biology, pharmacy, and chemical engineering (just to name a few!). It is anticipated that by the completion of this course, students will be familiar with all of the following topics and techniques: Safety in the laboratory Interpreting and following scientific directions Keeping a proper lab notebook Names and proper usage of lab instruments Understanding of general properties of compounds (including solubility, miscibility, acid/base chemistry, etc.) Proper usage of glassware Isolation and purification techniques (including filtration, solvent removal, drying solutions, distillations, chromatography (thin-layer, column, and gas) and crystallization/recrystallization) Characterization techniques including spectroscopy and melting point determination Interpretation of scientific results including percent yield and recovery, melting point, boiling point, IR and NMR spectra, and Rf values Required Materials: A laboratory notebook with carbon(less) pages Approved safety goggles Lab coats Lab manual will be posted through Blackboard Typical text: C.F. Wilcox, M.F. Wilcox, "Experimental Organic Chemistry, A Small-Scale Approach", (3rd edition, 2010). Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Lectures --> Lecture sessions are designed to clarify the concepts covered in the lab, as well as give an overview of techniques that will be used in the lab. Attendance is expected: The labs are only 3 hours in duration, so these lectures will be where you learn everything that you’ll need. Lab exercises will be available on Blackboard for each week. Please be considerate of your fellow students during the lecture period. Disruptions of any kind will not be tolerated and may result in expulsion from the classroom. Laboratory --> You will be required to have appropriate clothing before being allowed to enter the lab. Pre-labs are due at the beginning of the lab, and results and postlabs are due at the beginning of the lab 1 week after completion of the experiment! You will be expected to adhere to all of the lab safety rules. You are all expected to do your part to maintain a clean lab environment as part of GLP (Good Lab Practices): All reagent and solvent bottles should be completely closed immediately after use; All spills and dribbles should be cleaned immediately; All glassware should be put away at the end of the lab, and walkways should be kept free of debris. The following is the distribution of possible points in the course: Library Searching Exercise Database Search Exercises (Spectroscopy and Chromatography) Lab Quizzes Reaction/Synthesis methods knowledge Appropriate choice of method Appropriate constituents and tools. Procedure/steps (summary and/or ordering) Stoichiometry problems Spectroscopy and/or Chromatography analysis/interpretation Applications and industries Multistep Reaction/Synthesis Labs Lab Cleanliness Pre-lab Submissions Lab Notebook and Reports Lab Final Day 1: Much resemblance to quizzes Day 2-3: Augmented with the following: Molecular modelling software exercises Two or Three Practicum Group Labs (open notes) Part A. Points deducted for incompetent questionnaire for safety procedures for respective lab Part B. 2-3 labs to be implemented with competent data recording and lab reports. YOUR LAB REPORT CONSISTS OF THREE (3) PARTS --> Part I - Prelab Report. A copy of your lab notebook pages containing the lab write-up and answers to any prelab questions. This is due at the start of each experiment. Part II - Results. A copy of your notebook pages containing observations noted during the lab experiment. Is due with Part III one week from the conclusion of the experiment. Part III - Postlab Report. A summary of results and answers to postlab questions. This can be written on separate loose-leaf paper. Is due with Part II one week from the conclusion of the experiment Course Outline: Week1 Check-in/Safety Video/ Safety Procedures and Regulations Fractional Distillation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation, results and analysis Week 2 Measuring the Melting Points of Compounds and Mixtures Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 3 Purification by Recrystallization and Melting Point Measurement Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 4 Nucleophilic Substitution: Synthesis (SN1 Mechanism and SN2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 5 Oxidation of Alcohols (Primary, Secondary and Tertiary). Infrared Spectroscopy. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Infrared Spectroscopy Results and analysis Week 6 Elimination Reaction (E1 Mechanism and E2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 7 Synthesis of Aspirin. Chromatography and/or Spectroscopy Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Chromatography and/or Spectroscopy Results and analysis Week 8 Solvent Extraction Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 9 Electrophilic Aromatic Substitution: Synthesis of o- and p-Nitrophenol. No distillation; extract product with ethyl acetate. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 10 Separation and purification of o- and p-Nitrophenol by Liquid Chromatography. Use 100 mg sample, check by chromatography. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Results and analysis Week 11 Aldol Condensation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 12 Grignard Reaction: Synthesis of Phenylmagnesium Bromide. Week 1: Part 1. Add methyl benzoate and sustain the desiccator for next week. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 13 HCl workup of previous week’s product. Synthesis of Triphenylmethanol and recrystallization of product. Purity check by melting point measurement. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 14 -15 Wrapping up/cleaning things up. Final Exam. Prerequisite: Organic Chemistry I
Biochemistry: The study of biochemistry investigates the interplay between biological macromolecules such as proteins and nucleic acids, and low molecular weight metabolites (such as the products of glucose metabolism). In this course, you will apply your knowledge of intermolecular forces, thermodynamics (when a reaction occurs), chemical kinetics (how fast a reaction occurs), and chemical structure and functionality to understand how biological molecules (and life) work. COURSE GOALS AND OBJECTIVES (Our Roadmap!) -Be able to describe/identify the forces that direct/stabilize different levels of protein structure -Be able to predict how changes in amino acid (or nucleotide) sequence can affect macromolecular structure and function -Be able to explain how enzymes are able to affect reaction rate enhancement -Be able to articulate and apply what the enzyme parameters of KM, Vmax, kcat and kcat/KM tell us about an enzyme -Be able to describe the interactions of biomolecules both quantitatively and qualitatively (in many cases, including mechanistic details) -Be able to understand the flow of metabolic intermediates through a pathway and communicate information about metabolic pathways using diagrams -Be able to describe multiple experimental methods used in biochemistry, interpret data from these methods to form conclusions, and develop a testable hypothesis to answer a question -Be able to summarize and analyse primary literature and data, and apply gathered information to new situations -Increase problem solving skills such as: critical thinking, data analysis, graphical analysis -Increase process skills such as: communication of scientific concepts and experimental results, group dynamics and teamwork, management and self-assessment -Develop a community of active learners who are intentional about their educational choices Course Materials: Calculator Emphasis on reinforcing skills with software --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Typical Texts --> Nelson DL and MM Cox. Lehninger Principles of Biochemistry (5th edition). (“Lehninger”) Loertscher J and V Minderhout. Foundations of Biochemistry (3rd edition). (“FOBC”) Additional --> Blast protein databases, align protein sequences, build protein homology models, and evaluate the quality of these models Lab Manual example --> Lasseter, B. F. (2020). Biochemistry in the Lab: A Manual for Undergraduates. CRC Press Related to week 6: Edwards, P., Zhang, C., Zhang, B. et al. Smartphone based optical spectrometer for diffusive reflectance spectroscopic measurement of hemoglobin. Sci Rep 7, 12224 (2017). Course Overview --> You will frequently be given initial assignments to work on as an individual before class. These assignments must be ready at the start of class – your preparation will form part of your weekly participation grade. During our class meeting time, you will frequently function as a member of a Learning Team, developing and examining chemistry concepts as a unit. Your team effort and participation is part of your weekly participation grade. The team responses to a few Key Questions on each in-class activity will be evaluated for strength of concept and effective communication of the concept. The team will also strategize on ways to improve teamwork and team products. These responses will also form part of your weekly participation grade. Application exercises will be assigned for each activity. Together with problems from the text, they will form your weekly problem set that will be collected and graded for each individual. These homework problems and exercises are important to your success in the course. Actively working these homework problems is essential for your understanding of the material, as they bring your concept development full circle. The questions will be drawn from lectures, in-class activities, problem sets and discussions, as well as relevant primary literature that you may not have been previously assigned. The purpose of doing biochemistry is to gain experience in experimental methods that you’ll be reading about throughout the semester. Attendance on your scheduled lab day is expected. Software activities concerning biochemistry will accompany labs. Software activities concerning biochemistry will accompany labs as pre-lab development or post simulations. Grading --> Team Participation Problem Sets/Other Laboratory 2 Midterm Exams Final Exam Lecture Outline --> Week 1 Introduction to Biochemistry Week 2 Intermolecular forces and water. Amino acids and peptide bonds Week 3 Protein Folding Week 4 Working with proteins Week 5 Enzyme catalysis. Enzyme Kinetics Week 6 Enzyme inhibition. Hemoglobin Week 7 Exam 1; Carbohydrates Week 8 Glycobiology Week 9 Lipids and membranes. Transport across membranes Week 10 Signal transduction. Metabolism overview Week 11 Glycolysis. Glycolysis regulation and related pathways Week 12 Glycogen metabolism and gluconeogenesis. Citric Acid Cycle Week 13 Electron Transport Chain / Oxidative Phosphorylation; Exam 2 Week 14 Lipid metabolism. Nucleotides and nucleic acids Week 15 Nucleic acids structure and function Week 16 Final Exam Prereqs: General Biology I. Co-requisite or Prerequisite: Organic Chemistry I Cell Biology: The standard definition of a cell in most introductory biology texts includes the line that cells are “the fundamental building blocks of all organisms.” Because of this fact, trite though it may be, a detailed understanding of the fundamental processes of cellular function is critical to all specialties within biology, clinical or academic. Some of these processes, including for example the biochemical mechanisms underlying cellular energetics, are remarkably consistent from bacteria to human. Other cellular processes and structures vary from cell type to cell type or organism to organism, allowing for unique adaptations of cells and organisms to particular functions. For example, nerve cells have various properties allowing them to conduct electrical signals and therefore process information, while kidney cells are specialized for the secretion of waste, and red blood cells for the transport of oxygen and carbon dioxide. What are the differences in physiology from cell type to cell type determining these specific functions? During the first half of the semester we will focus primarily on the biochemical processes that underlie cellular function, with an emphasis on protein structure and function, ion transport mechanisms and energy metabolism. The second half of the semester will emphasize more the function of particular organelles, including cell membranes, intracellular compartments and the cytoskeleton, and the relevance of these structures on processes like cell signaling and mitosis. Throughout the course, we will emphasize how variability in these processes imbues different cell types with their unique functional abilities. We will also seek to understand the experimental evidence for the different facts and concepts we study: How do we KNOW that nerve cell signaling, for example, involves the release of neurotransmitters? Some of this experimental evidence will be explored in a hands-on way in the lab sections, some will be discussed during lecture, and some will be the subject of analysis in the reading of original scientific manuscripts. Finally, we will examine how malfunctions in the cellular processes we are studying underlie certain diseases. In particular, the final few lectures of the course will focus on the biology of cancer cells: how do changes in cellular processes allow cancer cells to proliferate and metastasize? What are some of the current clinical approaches to curing cancer by blocking or reversing these processes? Aspirations --> To understand fundamental concepts of cellular function. To understand, and be able to critically analyze, the scientific evidence underlying our current understanding of cellular processes. To develop skills, through lab experiments, in some of the specific methodologies used in the study of modern cell biology. To become skilled at formulating and testing hypotheses using these methods. To develop a preliminary ability to read and analyze the primary scientific literature: What are the major findings of a science paper? What evidence is presented to support these findings? Are there shortcomings, either in the methods used or the logic of the experiments, which might lead one to question the conclusions reached by the authors? To be able to put this knowledge into larger contexts of how disease states occur or how organisms function adaptively within their environments. Typical text: The World of the Cell, by Becker, Kleinsmith, and Hardin, 6th edition (2006), Pearson/Benjamin Cummings Class Requirements and Grading --> 1. Class Participation (10%) To include attendance, responses to questions I pose in class, participation in discussions, and simply raising your hand from time to time to ask questions or make a comment (something I DO expect you to do). 2. Quizzes (10%) Two short in-class quizzes during the first half of the course. 3. Homework/problem sets (5%) There won’t be many of these; I’ll assign them when we hit subjects that are especially involved to help you learn the material and to make sure everyone is on track. 4. Primary literature readings (10%) We will read two papers from the primary scientific literature during the second half of the course. In both cases there will be an in-class discussion of the paper and a “reading guide” set of short essay questions which will be graded. The first reading guide assignment will be due AFTER the in-class discussion; the second assignment will be due BEFORE the in-class discussion. 5. Laboratory reports (25%) The specifics of each week’s lab report will be discussed during lab section. Typically, each week’s lab report will be due the following Monday in lecture. 6. Mid-term exam (20%) TENTATIVE format to include an in-class component, a short oral component, and a take-home component. 7. Final exam (20%) LABS --> Lab instructions for each week will be handed out ahead of time, either distributed as hard copies in lecture or posted on the course Angel site (or both). You are responsible for reading the instructions before lab. Otherwise, labs tend to run late, you will have difficulty obtaining the necessary data and knowing what to do with it. Do not expect the instructor to go over every step of the lab procedure before you start. Labs will make great emphasis on strong, practical and constructive immersion into the following software to accompany hands-on activity: << VCell, TiQuant + TiConstruct + TISIM >> Such software provides strong quantitative/computational microscopic assessment of specimens (or whatever) at professional standards. Such provides better means of objectives and expectations towards hands-on labs. Each lab will be associated with an explicit lab report assignment (contained in the lab instructions), usually due in lecture the Monday following lab. Usually, you may either submit the report with your lab partner or independently. If a report is submitted jointly, both partners must have contributed equally, as per Honor Code responsibilities. Do NOT make the mistake of dashing off reports the night before in a single draft. These reports will collectively account for 25% of your course grade, so take them seriously. The lab is a potentially dangerous place and you are required to follow all instructions given by your lab instructor and presented in the lab instructions. Disregarding instructions, or coming to class late or unprepared, may result in grade penalties, in addition to being just plain dangerous for yourself and those around you. Note: students can apply stationary video recording of labs with assigned regulations (to be given). Course Topics: Chapter 1 -19, 24. Some topics will require at least one week of instruction. Labs --> Cell Culturing, Aseptic Technique Cell Culture: basic techniques, population curve Cell Counting and splitting plates Cell Staining Histology Electron microscope Cell Harvesting & Cell Lysis Fractionalization of cells Common method(s) will be implemented Discussion and logistics for immunomagnetc separation & magnetic beads Isolation of erythrocyte membrane proteins Analysis of erythrocyte membrane proteins Bradford Assay (also identifying advantages and disadvantages) SDS-PAGE Chloroplasts and the Hill reaction Prerequisites: General Biology I & II
Microbiology I Microbiology is an integral part of many different scientific studies, such as immunology, genetics, molecular biology, biochemistry, medicine, agriculture, ecology, industrial processes and many more. People working in these fields use microbiology in their daily procedures, although they aren't microbiologists. Because of the wide range of its applications, understanding the basics of microbiology is in many ways essential to our completeness as biologists, no matter what field we may pursue. Microorganisms (in the context of this course) are minute living things that are individually too small to be seen with the naked eye. The term includes bacteria, microscopic fungi (yeasts and molds), protozoans, microscopic algae, prions and viruses. Microorganisms can be associated with many diseases, infections and inconveniences such as AIDS, pimples, and spoiled food. However, the majority of microorganisms make vital contributions to the world's inhabitants. They maintain the balance of chemicals and living organisms in the global environment. For example, the algae and cyanobacteria found in the oceans and waters of the globe are the major source of oxygen for living things. In many places microorganisms are the basis for the food chain. They help to recycle chemical elements in the land and water. Microorganisms also have been used for commercial benefits. Cultured microorganisms can be used to synthesize products more cheaply than they can be manufactured by other means (biotechnology). Microorganisms have also been used to produce products that have "always" been a part of our lives, such as vinegar, wine, sauerkraut, pickles, beer, green olives, soy sauce, buttermilk bread, cheese, and yoghurt, to name a few. The purpose of this semester of Microbiology is to familiarize the student with those concepts that are basic to viruses and prokaryotic and eukaryotic cells. Lecture is the foundation of the course. Laboratories will not always coincide with the lecture topics, as the laboratories are designed to give the student the basic laboratory techniques necessary to identify microorganisms. The student is responsible for assignments (such as designated papers from the scientific literature) that add to the lecture and lab material. In order to enhance appreciation of the course, the student is encouraged to seek out related materials that are available, such as scientific journals, JoVE, etc.. There are five basic topics in this course - the general principles for microbial the growth, evolution and classification; descriptions of different prokaryotic, eukaryotic and other lifeforms and how they utilize these principles; the natural ecology of microorganisms; the human use of microorganisms; and how microorganisms function in disease. Section one covers the first topic, the second topic is covered by sections two and three, and the final three are covered in section four. Some aspects of the last two topics are woven throughout the course. In order to understand how microorganisms can live, one must know what the parameters for their existence are. In order to get a feel for the diversity and scope of the microbial world, one must have a feel for what kinds of organisms exist, and to understand the ecology, uses and dangers of microorganisms, one should have a general knowledge of the different organisms to be encountered. All of these things are useful in life, in order to make informed decisions, and to go on to professional or graduate school. Learning activities will include reading and evaluation scientific papers, learning basic Microbiological techniques, identifying unknown bacteria, answering questions in lecture and writing scientific papers. Learning Activities --> These will consist of lectures, laboratory demonstrations, laboratory work (including independent investigation to identify unknown organisms), reading assigned scientific papers, writing a final laboratory report and answering those questions that are asked in lecture and laboratory. Outside the formal lecture/ laboratory structure, the student is expected to read assignments in the text, as well as assigned papers from the scientific literature, and study the concepts presented in lecture, laboratory and in the text. Hopefully this mix of learning styles will create a deeper appreciation of Microbiology. Typical texts in Unison: Microbiology; Prescott, Harley and Klein, 6 ed. (LECTURE) Microbiology in Practice ed.6; Beishir (LAB MANUAL) Grading: 3 Exams 45% Labs 25% Final Exam 30% MATERIALS --> Standard Operating Procedure for Handling Laboratory Specimens (SOPFHLS) and so forth. All specimens will be handled using the "Standard precautions for blood and body fluids" recommended by public health (CDC) officials and government agencies. Practice will include: 1. Specimen brought into the MLT Laboratory will be carefully screened, in proper containers and labeled. 2. Proper protection will be used when processing specimen (lab coats, gloves and protective eye wear as appropriate). 3. Mechanical pipetting, use of safety engineering devices, and safe work practices at all times. 4. Decontamination of laboratory work surfaces at end of each exercise. Use bleach and/or phenol. 5. Decontamination or proper disposal of contaminated laboratory test materials. a. Blood and body fluids will be placed in biohazard bag in the trash. b. All needles, broken glass, and hemolets will be placed in a biohazardous sharps container and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. c. All gauze, Kim wipes and disposable materials contaminated with blood and body fluids will be discarded in a biohazard bag and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. 6. Decontamination of scientific equipment, i.e., electrodes, glassware, etc. 7. Hand washing after all laboratory procedures. 8. Accurate and proper recording and reporting of results. Reference: "Legal Implications of Universal Blood and Body Fluids Precautions", David L. Wing, Clinical Laboratory Science, Volume 1, No. 2, March/April 1988. " Needlestick Safety Act", OSHA Standards 2002 (or later version) 9. << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> 10. A standard academic laboratory manual accompanied by the likes of the following << https://www.mountsinai.on.ca/education/staff-professionals/microbiology >> 11. Microbial, Viral and pathogen software (check Goody Bag post) will follow applicable and practical mathematical modelling of cultures and so forth w.r.t. to median, nutrition and other environmental factors. All such along with lab studies of cultures --> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> LABORATORY EQUIPMENT --> Equipment: PPE-impervious lab coat, latex or nitrile gloves, closed toed leather shoes PERIODICALS--> 1. American Journal for Medical Technology 2. Laboratory Medicine 3. Medical Laboratory Observer (MLO) 4. Journal of Clinical Microbiology RESOURCE --> << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> LABS --> The Laboratory is set up to familiarize the student with the techniques necessary to grow and identify microorganisms. The first quarter of the semester covers basic techniques of media preparation, staining and microscopy. The second quarter covers recognition and differentiation of microbial characteristics in culture. The next section is devoted to microbial identification based on metabolic differences. The student will then test his or her knowledge by using the above information to identify a mixture of two unknowns in the last section of the course. A six page research report of the unknown identification will be required, in formal scientific paper format (including bibliography), and will count for 20% of the total grade in the course. COURSE TOPICS --> SECTION ONE: The Basics of Microbial Existence and Detection Introduction – Chapter 1 Microscopy – Chapter 2 Prokaryotic Structure & Function – Chapter 3 Microbial Nutrition – Chapter 5 Microbial Growth – Chapter 6 Control of Microbes – Chapter 7 Taxonomy – Chapter 19 EXAM 1 SECTION TWO: The Bacteria Gram Negative Bacteria – Chapters 21 – 24, 20 EXAM 2: from taxonomy through the Archaea SECTION THREE: The Eukaryotes and Viruses Eukaryotic structure & Function – Chapter 4 Fungi – Chapter 25 Algae – Chapter 26 Protista – Chapter 27 Viruses – Chapter 16 Prokaryotic Viruses – Chapter 17 Eukaryotic Viruses – Chapter 18, 38 EXAM 3: from Eukaryotic Structure & Function through the viruses SECTION FOUR: Microbial Ecology; food, industrial and medical microbiology Symbiosis – Chapter 28 Aquatic Ecology – Chapter 29 Terrestrial Ecology – Chapter 30 Industrial Microbiology – Chapter 42 Food Microbiology – Chapter 41 Medical Microbiology – Chapter 34 – 37; 39 – 40 FINAL EXAM LABS TO BE DONE -- > Laboratory Safety, Microscopy, Aseptic Technique – Modules 4, 5, 6 Bacterial Cultures, Slide Preparation, Staining, Streaking – Modules 22-24 &12 Streaking, Pour Plates – Modules 7, 8, 9, 10, 12 Bacterial Characteristics – Modules 11, 12, 13, 25 Differential and Selective Media, IMViC Test – Modules 39, 40, 49, 50 Bacterial Identification – Modules 34, 37 Litmus Milk, Carbohydrate Fermentation, Hydrogen Sulfide, Agglutination Tests – Modules 34, 37 Further Tests – Modules 32, 35, 36, 38, 46 Identification of Unknowns – 56, all of the above Hašek J. (2006) Yeast Fluorescence Microscopy. In: Xiao W. (eds) Yeast Protocol. Methods in Molecular Biology, vol 313. Humana Press Prerequisites: General Biology I & II, General Chemistry I & Ii Microbiology II Applying microbiological techniques to study and identify unknown organisms and the use of molecular genetic techniques to study and manipulate microbes. Lecture material will provide the theory and background to the laboratory exercises done each week. The students will collect data from laboratory exercises and incorporate their methods and results into a research report that follows the format of research manuscript. STUDENT INTELLIGENCE --> Recognising how biochemical and genetic tests are used to identify unknown organisms. Comprehending how DNA sequencing can be used to identify microbes. Describing useful components of plasmid cloning and expression vectors. Learning the theory of PCR, how PCR primers are designed, and how to add desirable sequences to PCR products using primer modifications. Developing written communication skills by writing experimental results in manuscript form PRACTICAL SKILLS --> Develop the ability to store and maintain microbial cultures. Determine the appropriate tests to use for the identification of an unknown organism. Use genetic techniques to study and modify microbes. Clearly and concisely communicate research results. Use computer programs for bioinformatics analysis. MATERIALS --> Standard Operating Procedure for Handling Laboratory Specimens (SOPFHLS) and so forth. All specimens will be handled using the "Standard precautions for blood and body fluids" recommended by public health (CDC) officials and government agencies. Practice will include: 1. Specimen brought into the MLT Laboratory will be carefully screened, in proper containers and labeled. 2. Proper protection will be used when processing specimen (lab coats, gloves and protective eye wear as appropriate). 3. Mechanical pipetting, use of safety engineering devices, and safe work practices at all times. 4. Decontamination of laboratory work surfaces at end of each exercise. Use bleach and/or phenol. 5. Decontamination or proper disposal of contaminated laboratory test materials. a. Blood and body fluids will be placed in biohazard bag in the trash. b. All needles, broken glass, and hemolets will be placed in a biohazardous sharps container and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. c. All gauze, Kim wipes and disposable materials contaminated with blood and body fluids will be discarded in a biohazard bag and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. 6. Decontamination of scientific equipment, i.e., electrodes, glassware, etc. 7. Hand washing after all laboratory procedures. 8. Accurate and proper recording and reporting of results. Reference: "Legal Implications of Universal Blood and Body Fluids Precautions", David L. Wing, Clinical Laboratory Science, Volume 1, No. 2, March/April 1988. " Needlestick Safety Act", OSHA Standards 2002 (or later version) 9. << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> 10. A standard academic laboratory manual accompanied by the likes of the following << https://www.mountsinai.on.ca/education/staff-professionals/microbiology >> 11. Microbial, Viral and pathogen software (check Goody Bag post) will follow applicable and practical mathematical modelling of cultures and so forth w.r.t. to median, nutrition and other environmental factors. All such along with lab studies of cultures --> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> ASSESSMENT --> 3-4 Quizzes Labs (activity + lab reports) 3-4 Exams COURSE OUTLINE --> WEEK 1 Introduction to cloning Vector analysis; Computer programs; Start cultures Plasmid preps and DNA quantification WEEK 2 PCR and primer design; Primer design programs Using primer design programs WEEK 3 PCR theory; Preparing a PCR Set up PCR PCR gel electrophoresis WEEK 4 Discuss purification and quantification of DNA Purify PCR Quantify PCR; Discuss restriction enzymes and protocols WEEK 5 Restriction digestion set up (PCR and plasmid) Inactivate digests; Discuss ligation Purify digests, quantify, and perform ligation reactions WEEK 6 Discuss E. coli strains and transformation Transformation protocol E. coli transformation and plating WEEK 7 How to analyze and interpret culture results Set up colony PCR; start cultures Purify plasmids; Colony PCR and plasmid electrophoresis WEEK 8 Cryopreservation Start cultures for cryopreservation Cryopreserve cultures; Discuss conjugation protocol WEEK 9 Finalize conjugation protocol; start SM10 and E. ictaluri cultures Mix cultures, filter, and plate Resuspend conjugation, dilute, and plate WEEK 10 Analyze conjugation results and discuss analysis Set up colony PCR and start cultures Colony PCR electrophoresis; Plasmid preps; Cryopreserve WEEK 11 Project Introduction; Sampling General Techniques Refresher Choose colony of interest; Staining, motility, and other tests WEEK 12 Streak fresh cultures Start broth culture Cryopreserve; Genomic DNA purification WEEK 13 Quantify gDNA; discuss 16S PCR Set up 16S PCR Apr 28 PCR electrophoresis; PCR purification WEEK 14 Using biochemical analyses and identification systems Inoculate biochem media and Biolog Biochem and Biolog Interpretations WEEK 15 Discuss final analyses Repeat biochems or perform additional tests Interpret final tests Prerequisites: Microbiology I Genetic Engineering & Technology Course textbook: Molecular Biotechnology: principles and applications of recombinant DNA, by B.R. Glick and J.J. Pasternack Course Grade Constitution --> 3 - 6 Assignments 3 Exams Course Outline --> Week 1 Introduction-Genetic Engineering and a tour of Genome Space DNA/RNA Processing and Gene Expression (a review): Week 2 Basic Techniques of Molecular Biology Restriction Endonucleases, Vectors, Cloning Week 3 Library Screens PCR, Sequencing Week 4 Exam Review and Exam Week 5 Prokaryotic Gene Expression Eukaryotic Gene Expression Genetic Engineering in Plants I Week 6 Genetic Engineering in Plants II: Applications Genetic Engineering in Plants III: the Next Generation of Rice Week 7 Sequence Analysis, Genome Structure Comparative Genomics Week 8 Functional Genomics: Analysis of Gene Expression Modifying Gene Expression and Cellular Function Week 9 Exam review and Exam Week 10 OPV and the emergence of AIDS The Origin of AIDS, con’t; vaccine intro; Edible vaccines Vaccine targets: malaria and ebola Week 11 Human Gene therapy I Human Gene therapy II: examples Week 12 Genetic engineering in animals I: Cloning Genetic engineering in animals II: Knockouts and Knockins, Inducible Gene Targeting Week 13 Genetic engineering in animals III: examples Genetic engineering in animals IV: xeno transplantation part a Genetic engineering in animals IV: xeno transplantation part b Week 14 Ethics and Patent Law Week 15 Exam Review and Exam Prerequisites: Cell biology, General Chemistry II Molecular Biology I This Molecular Biology course is designed to give a good background in current Molecular Biology, which should allow for easy continuation to graduate or professional school courses. The major themes are Eukaryotic and Prokaryotic DNA replication, Chromosomal structure and function and Gene structure and function. Students will learn from current papers in the scientific literature, and will be expected to use concepts developed in the course in class, in the laboratory and in exams. Molecular Biology is fundamental to the study of all living things. It describes, in its most basic form, the mechanisms of how organisms live, reproduce and evolve. It is basic to much of modern Biology, no matter what the field of study. The purpose of this semester in Molecular Biology is to familiarize the student with those concepts that are basic to the functioning of prokaryotic and eukaryotic cells. Lecture is the foundation of the course. Laboratories will not always coincide with the lecture topics. The student is responsible for assignments that add to the lecture and lab material. The student is encouraged to seek out related materials that are available, such as scientific journals (e.g. Cell, Nature, Scientific American), newspapers, magazines and television programs (e.g. channels 12 and 52) that relate to course topics. Lecture notes will be published on the internet at my home page before the given lecture. There are three basic concepts in this course - the replication of DNA; the structure and function of chromosomes; and the structure and functioning of genes. They will be organized as indicated below. Students will reinforced with Molecular Biology laboratory techniques (DNA isolation and purification, recombinant DNA synthesis and cloning, gene detection, PCR and Southern and Western Blotting), which will be used to expand the student's appreciation and knowledge of the lecture material. The lectures cover Molecular Biology as a whole - the "central dogma" of Biology: DNA makes RNA which then makes protein. Out of this there arise three concepts - Eukaryotic and Prokaryotic DNA biosynthesis; Chromosomal structure and function (with associated proteins and functions); and Eukaryotic and Prokaryotic gene structure and function (mRNA, tRNA synthesis and function, including protein synthesis), and how they relate to basic biological and chemical concepts (such as the action of evolutionary processes on living things) learned in previous courses. In general, they should understand how our genomes function, including gene activation and deactivation, RNA synthesis and protein biosynthesis and be able to use this knowledge in their work and in the laboratory. Overall, emphasized and reemphasized in the course, and illustrated by specific examples and laboratory experiments, are the ways in which the above topics are interconnected, and factors used in one way are recycled to be used in another. This leads to interconnectiveness amongst the various cellular functions, and allows for signaling and controls between them. These principles should allow them to establish a firm connection between this course and other aspects of biology and give a foundation for future Molecular Biology courses and/or a good appreciation of concepts needed to make reasoned choices in their everyday lives. Typical Text: Watson et. al. Molecular Biology of the Gene ed. 5 Typical Laboratory Manual Text: Human Molecular Biology Laboratory Manual - S. Surzycki The professor's evaluation of student participation in lecture and laboratory can be used to benefit hard working students and possibly enhance their grade if they are in a borderline position. The laboratory grade is based mainly on the laboratory paper (normal scientific format, aprox. 8 - 10 pages, with a bibliography and internal referencing), as well as the instructor's assessment of the student's activity for the entire laboratory. Some laboratory based questions will appear on exams, especially including the final exam. Grading: Exams -15% of final grade (x 3 exams): 45% Laboratory -25% of final grade Final Exam -30% of final grade Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well in this course that further encourages a modern and profession environment, extending beyond memory based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities of interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Lab Outline --> Chapter 7 - Determination of Human Telomere Length (pages 164 - 195) All activities in chapter 7 must be done Chapter 8 - RT-PCR of Human Genes (pages 196 - 214) All activities in chapter 8 must be done Course Outline --> SECTION ONE: DNA replication, repair and recombination Chapters 1 – 11 SECTION TWO: Chromosome structure and function, chromatin, prokaryotic operon structure and function Chapters 7 & 16 SECTION THREE: The eukaryotic operon structure and function, gene clusters, genes in organelle Chapters 2, 3, 17 & 18 SECTION FOUR: Ribosomes, protein biosynthesis and transportation, eukaryotic and prokaryotic viruses, genetic engineering Chapters 14 & 16, 17, 13 LAB REPORT DUE & FINAL EXAM Prerequisites: Cell Biology, Organic Chemistry I
Molecular Biology II Molecular Biology is a wide term encompassing two often complementary fields of study: a) laboratory and computer-based tools that can be used to study gene and genome identity and function (“molecular tools”), and b) the underlying fundamental structure of DNA and RNA. In this class there will be about a 50:50 split between focus on molecular tools (techniques) on the one hand, and the structure and function of DNA and RNA on the other hand. Proteins are often the focus of Biochemistry classes. Molecular Biology stresses the application of advanced molecular tools in the lab, and during analysis of scientific data presented in primary literature, in addition to covering genetic topics in more detail than was done in previous classes. The major expectations of students are : • To be become familiar with molecular techniques. • Understand the use of these techniques in the discovery of DNA and RNA metabolism and function. • Become more proficient at reading and critiquing primary literature. • Become familiar with commonly used laboratory techniques in the lab during an allsemester long research project. • Emphasis is not on memorization of the details of the molecular machinery of the cell. Instead, it is on developing skills to apply the learned techniques to the understanding of scientific discovery (data interpretation), as well as to suggest ways to study the function of molecules (experimental design). Molecular research is impossible to conduct in set 4-hour increments once a week. Typical Textbook/Readings: • Burton Tropp: Molecular Biology, 4th edition, 2012 Parts of some chapters will be used in this book in lecture, and book will be a good resource for looking up details, reading ahead or after class. Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well to this course that further encourages a modern and profession environment, extending beyond memory based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities of interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Grading --> 2 Midterm exams (each 70 points) 140 points 5 Quizzes (4x10 points each, quiz 5 =20 points) 60 points 1 Research paper (lab write-up) 40 points Lecture participation 20 points Carefulness in lab and preparation for labs 20 points Lab prelabs/postlabs ~30 points 3 Paper discussion prep and participation (4x20 points + 2x10 points) 100 points LABS --> It’s important to keep a neat and bound notebook for the lab, but you do not need to buy an expensive lab notebook. At the end of the semester you will be required to turn in a research paper describing the work you did on the lab project and put it in context of a greater scientific question. Only one lab report/paper will be handed in for the semester. This report reflects multiple weeks of work. Therefore make sure to collect gel photos, instrument readings, microarray and RNA-seq data analyses, so that you have all the files you need for your final paper. While all class and lab assignments have to be written individually unless specified differently, the final lab report can be written collaboratively with your lab partner, or individually if you prefer. Preparation for labs: Labs in this class are very expensive and it is of great importance that you come prepared to lab. In the past I have simply relied on suggesting that everyone read the manual before lab. However, it only takes one un- or underprepared person to make an experiment fail – often for more students than just that one person. So I have decided that some form of enforcement of the preparation requirement is needed and have reverted to prelab assignments, which test your level of preparedness. In addition there is now a grade for “carefeulness in lab”. This grade is based on whether you are prepared for lab or excessively ask unnecessary questions that you could have answered yourself by reading the manual. I do encourage asking questions, but I also encourage self reliance and careful attention to detail. Not every experimental failure is due to operator error. Such failed experiments are common and will not influence your lab grade negatively. Again, software mentioned earlier will accompany labs. Outline Week 1: Nucleic Acid Structure, Genome Organisation Week 2: RNA techniques (hybridization, reporters) RNA techniques (qPCR) DNA sequencing methods (Sanger) DNA sequencing methods (whole genome approach) Lab1: Microarray analysis - RNA Extraction Week 3: QUIZ 1: RNA techniques DNA sequencing methods (whole genome approach) Paper Discussion 1 Lab1: Microarray analysis c - DNA production Week 4: Gene mapping, map based cloning Human genome variation, the concept of “race” Lab1: Microarray analysis - Slide hybridization Week 5 Paper Discussion 2 - Human genome variation DNA Damage Quiz 3 (mapping/cloning) Lab1: Microarray analysis Statistics of microarray analysis Slide analysis: first part in lab. Finish slide analysis/statistics on your own time after all slides are pre-analyzed after [whenever] Week 6: DNA repair, Technique: EMSA (for paper 1) Paper Discussion 3 (DNA repair) Lab 2: RNA-seq analysis - Computer workshop using iPlant tools (or by software provided) Week 7: Recombination Lab 2: RNA-seq analysis - Analysis of RNA-seq data from week 1, and comparison with array data Week 8: Transposons Paper Discussion 4 (Recombination) Lab 3: Genetic marker analysis Sequence analysis using Genome Browser Primer design CAPS search Week 9: Eukaryotic transcriptional regulation Lab 3: Genetic marker analysis - DNA extraction, PCR set up Week 10: Epigenetics (technique: Immunoprecipitation) Paper Discussion 5 (Epigenetics ) Lab 3: Genetic marker analysis CAPS digest gel analysis (on your own) Lab 4: RNAi cloning - PCR of insert Week 11: QUIZ 4: The transcription unit RNAi Lab 4: RNAi cloning PCR clean-up and cloning reaction Transformation (on your own) Week 12: Paper Discussion 6 (RNAi) Lab 4: RNAi cloning - Finish up from week 11 Week 13: Splicing Agrobacterium and gene transformation Lab 4: RNAi cloning Colony PCR of cloned insert Glycerol stocks and sequencing of positive clones Week 14: Agrobacterium and gene transformation QUIZ 5: (molecular techniques) Lab 4: RNAi cloning Finish up from week 13 Final Exam Lab report due Prerequisites: Genetic Engineering & Technology, Molecular Biology I Biotechnology Laboratory: This course covers the basics and of laboratory and analytical techniques that are used in biomedical research and the biotechnology industry. The techniques are in the following areas: (1) biochemistry of bacterial recombinant proteins, (2) mammalian cell culture, (3) molecular and cell biology, and (4) mass spectrometry. This is a laboratory course and serves as a pre-requisite for Advanced Biotechnology Laboratory. Specific topics include: general laboratory safety, record keeping, preparation of research reports, manipulation of bacteria, protein overexpression and purification, enzyme assays, highthroughput techniques, high performance liquid chromatography (HPLC) and mass spectrometry, mammalian cell culture, Western blotting, protein-protein interactions, reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and assays for gene expression. Learning Objectives: -Acquire basic knowledge in protein biochemistry and bacterial molecular biology and experimentation with common equipment in a laboratory environment. -Gain basic knowledge in the use of mass spectrometry (MS), high performance liquid chromatography (HPLC), and automation in a laboratory environment. -Gain basic knowledge in mammalian cell culture and mammalian molecular and cell biology experimentation with necessary equipment in a laboratory environment. Prototypical Textbook: Basic Laboratory Methods for Biotechnology (Seidman & Moore) and additional materials. ACCOMPANIED BY LAB MANUAL(S). Assessment --> Problem Sets 10% 3 Lab Quizzes 15% Labs 45% 2 Lab Exams: mid-term (15%) and final (15%). Problem sets (25%): two problem sets will be given and are due at midnight given dates at midnight, respectively. Answers to problem sets are written concisely and clearly. Group Presentations for (15%): each student will give a 15-20 min. Oral presentation based on published articles. Course Outline: WEEK 1 Overview of modern biotechnology successes Basic experimental facets (basic instruments, buffers/solutions) I WEEK 2 Basic experimental facets (basic instruments, buffers/solutions) II Mammalian cell culture: equipment, starting cell culture, culturing and freezing cells WEEK 3 Molecular cloning: Restriction enzymes, plasmids and intro to cDNA and genomic libraries Detecting nucleic acids: agarose gel electrophoresis, PCR and qPCR, Southern/Northern blotting WEEK 4 Studying proteins (I): protein preparation, fractionation, SDSPAGE/Western blotting Studying proteins (II): protein engineering - plasmids and epitope tags and site-directed mutagenesis WEEK 5 Studying protein (III): detecting protein subcellular localization, immunofluorescence microscopy Transient transfection: endogenous vs. exogenous, principle and purpose WEEK 6 Studying protein-protein and protein-chromatin interactions: coimmunoprecipitation, GST pulldown and ChIP assays Reporter assays and qRT-PCR: studying transcription factors, protein function and drug screening WEEK 7 Knockout/knockin technologies Case study: Studying nuclear hormone receptors WEEK 8 In-class literature presentation (for ½ the group) Mid-term exam WEEK 9 Protein quantification and purity analysis WEEK 10 Protein expression in E. coli Purification of proteins by affinity tags and standard methods WEEK 11 Activity assay development Example of activity assay: β-lactamase WEEK 12 Biophysical characterization of protein-ligand interactions Fundamental aspects of mass spectrometry WEEK 13 Examples of analysis of mass spectrometry WEEK 14 Standard Operating Procedures (SOP) and record keeping in academia and industry Statistical analysis of data WEEK 15 How to prepare research reports In-class literature presentation: for other ½ of the group Prerequisites: Genetic Engineering & Technology, Molecular Biology I Advanced Biotechnology Laboratory: This course provides advance extensive hands-on laboratory experience in bacterial recombinant protein biochemistry, mammalian cell culture, molecular and cell biology, and mass spectrometry. Specific topics include: General laboratory safety, good laboratory practices (GLP), standard operating procedures (SOPs), buffers, media, and other reagent preparation, sterile technique, manipulation of bacterial and mammalian cells, mammalian cell culture, work with DNA and RNA, polymerase chain reaction (PCR) techniques including quantitative reverse transcription (RT-qPCR) and molecular cloning, protein overexpression and purification, assays (enzyme, stability, and reporter), highthroughput techniques, transfection, immunoprecipitation, immunofluorescence, DNA and protein gel electrophoresis, high performance liquid chromatography (HPLC), and mass spectrometry. Learning Objectives: -Acquire familiarity with and good laboratory practices (GLP), including use of standard operating procedures (SOPs). -Gain experience in protein biochemistry and bacterial molecular biology experimentation, including the necessary equipment in a laboratory environment. -Gain experience in mammalian cell culture and mammalian molecular and cell biology experimentation including the necessary equipment in a laboratory environment. -Gain experience in the use of mass spectrometry (MS), high performance liquid chromatography (HPLC), and automation in a laboratory environment. -Collaborate with peer scientists in a laboratory environment. -Prepare laboratory reports with an emphasis on procedures relevant to an industrial setting. This is a Monday to Friday laboratory course for 3 hours per day. Prototypical text: Basic Laboratory Methods for Biotechnology (Seidman and Moore) Required materials including laboratory protocols and standard operating procedures (SOPs) will be procured. There will be 2 exams: a lab practical exam and a written exam. The lab practical exam will be administered during the last week of classes in the laboratory space. The written exam will be administered during finals week. Laboratory notebook 30% Laboratory reports 30% Laboratory practical exam 30% Written final exam 10% Course Outline: PREMATURE FORMALITIES Biosafety training, autoclaving, sterile technique Calculations for buffers, solutions, and media Buffers, solutions, and media; autoclaving PROKARYOTIC TECHNIQUES -Nucleic acids and bacterial cell manipulation DNA quantification and polymerase chain reaction (PCR) Transformation and overnight cultures Plasmid preparation, sequencing, and restriction digestion DNA gel electrophoresis Data analysis -Recombinant protein expression in bacteria Sequence analysis; media preparation Transformation Culturing and overexpression Culturing and overexpression Spin down and collect cells -Protein purification Solution preparation Protein purification Size exclusion chromatography Size exclusion chromatography Protein gel electrophoresis and quantitation -Enzyme and stability assays Ultraviolet-visible (UV-vis) spectrophotometry assays UV-vis spectrophotometry assays Thermal shift assays Thermal shift assays Data analysis -Enzyme Assays 96-well plate assay preparation 96-well plate assay Data analysis Data analysis Data analysis and clean-up EUKARYOTIC TECHNIQUES -Maintenance of mammalian cells Making growth medium & thawing cells Making growth medium Splitting cells Making cell stocks Splitting cells -Transient transfection, immunoprecipitation, Western blotting Splitting cells, transient transfection Harvest cells & Immunoprecipitation Splitting cells, SDS-PAGE Western blotting Splitting cells -Transient transfection, immunostaining, immunofluorescence microscopy Split and seed cells Transient transfection Split cells, Immunostaining Imaging Split cells -Transient reporter assays Split cells, transient transfection Change media & add ligands Split and harvest cells & reporter assay Data analysis Split cells -Isolation of total RNA & RT-qPCR Treat cells with ligands Extract RNA from mammalian cells Reverse transcription (RT) of RNA to generate cDNA Quantitative real-time PCR (qPCR) Data analysis and clean-up MASS SPECTROMETRY -MS basics and familiarization Familiarization with mass spectrometry software Familiarization with high performance liquid chromatography (HPLC) software Tuning, standards, and optimization of direct infusion HPLC setup and operation: tuning, standards, and optimization HPLC operation -MS Assays Assays and HPLC/MS sample prep HPLC/MS runs HPLC/MS runs Data analysis Data analysis and clean-up -Wrap-up: Laboratory practical exam week Review No class; finish laboratory notebooks and laboratory reports Laboratory practical exam, part 1 Laboratory practical exam, part 2 -Last Day of Classes and Finals Week End-of-class clean-up and check-out; laboratory notebooks due Final written exam Prerequisite or Co-requisite: Techniques in Biotechnology
Environmental Microbiology: Course is highly dependent on knowledge, experience and from Microbiology I & II. Course will not be closely repetitive as prerequisite, rather, course concerns the development of competency, accuracy and efficiency for microbial observation, proper identification and removal. Apart from prerequisites reading and pre-study will be critical towards successful and efficient completion of lab obligations; outlines and other preparation documentation for a respective lab will be provided 5 to 7 days before day of respective lab administration. Instructor likely to provide a 15 – 30 minute synopsis for lab to done on day of relevance. Texts: TBA MATERIALS --> Standard Operating Procedure for Handling Laboratory Specimens (SOPFHLS) and so forth. All specimens will be handled using the "Standard precautions for blood and body fluids" recommended by public health (CDC) officials and government agencies. Practice will include: 1. Specimen brought into the MLT Laboratory will be carefully screened, in proper containers and labeled. 2. Proper protection will be used when processing specimen (lab coats, gloves and protective eye wear as appropriate). 3. Mechanical pipetting, use of safety engineering devices, and safe work practices at all times. 4. Decontamination of laboratory work surfaces at end of each exercise. Use bleach and/or phenol. 5. Decontamination or proper disposal of contaminated laboratory test materials. a. Blood and body fluids will be placed in biohazard bag in the trash. b. All needles, broken glass, and hemolets will be placed in a biohazardous sharps container and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. c. All gauze, Kim wipes and disposable materials contaminated with blood and body fluids will be discarded in a biohazard bag and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. 6. Decontamination of scientific equipment, i.e., electrodes, glassware, etc. 7. Hand washing after all laboratory procedures. 8. Accurate and proper recording and reporting of results. Reference: "Legal Implications of Universal Blood and Body Fluids Precautions", David L. Wing, Clinical Laboratory Science, Volume 1, No. 2, March/April 1988. " Needlestick Safety Act", OSHA Standards 2002 (or later version) 9. A standard academic laboratory manual accompanied by the likes of the following << https://www.mountsinai.on.ca/education/staff-professionals/microbiology >> 10. << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> 11. Microbial, Viral and pathogen software (check Goody Bag post) will follow applicable and practical mathematical modelling of cultures and so forth w.r.t. to median, nutrition and other environmental factors. All such along with lab studies of cultures --> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> Grade --> Attendance & Conduct 30% Lab Reports 30% Midterm 20% Final 20% Poor conduct in course and lab can warrant 69% percent of total grade for the course being forfeited. There are the conventional practices-- Observation & Removal Activities 1. Detection --Broth Culture, Quantal (“Most Probable Number”) Methods: Multiple Fermentation Tube (MFT) and Defined Substrate Methods and the Compartment Bag Test (CBT) --Enumerative (Colony Count) Methods: Membrane Filtration, Pour Plates and Spread Plates 2. Quantification --Quantification of Somatic and Male-specific Coliphages in Water and Wastewater 3. Microbial Survival in the Environment 4. Microbial Removals by Physical-Chemical Water Treatment Processes 5. Microbial Removals by Molecular Methods 6. Microbial Removal by Disinfection Processes, Chlorine and UV 7. Possible cases of excess, inappropriate or irresponsible implementation of removal methods, and the hazardous consequences. Note: (1), (2) and possibly (3) concern identification of target microorganisms that may vary from two or higher. Concerning topics (A) through (I) below there will be determination on how and when (1) through (3) are relevant, with the logistics, followed by experimentation procedures to confirm such. If (1) through (3) doesn’t serve appropriately then to determine which methods provide resolutions and orchestration of such lab activity; exams likely will demand such, which also may or may not depend on microorganism considered --> A. Bacterial Counts B. Water Content of Samples C. Contact Slide Assay D. Dehydrogenase Activity of Soils E. Nitrification and Denitrification F. Microbial Examination of Water G. Bacteriological Examination of Water H. Defined Substrate Technology I. Amplify 16S rRNA gene of isolates, verify via gel electrophoresis, and sequence J Biodegradation of Phenol Compounds Note: in lab reports sources of specimen must be identified and dated. Students must provide information on possible illnesses, defects or diseases related to a respective identified microorganism from respected sources and/or acquired intelligence from studies; incorporates describing the environmental parameter settings and vectors to trigger such. All such may also be asked on exams. An exam will not have questions from labs not given nor returned; all lab reports have deadlines regardless of whether students turn in lab reports or not. Each day a lab report is late warrants 5% reduction in respective lab report scoring. Prerequisites: Microbiology I & II Clinical Microbiology: The general characteristics of bacteria, protozoa, yeasts, molds, and viruses are used to understand the role of microorganisms in human health and disease. The interactions between the host and the microorganisms are emphasized as well as the physical and chemical methods of control. To develop procedural skills in Clinical Microbiology along with the thinking Processes that relate the following factors to each other --> Patient characteristics Types of infections Specimen requirements Microscopic examinations Culture media: growth characteristics and amounts Specific organisms and their identification techniques. Treatment and procedures relating to that treatment. Such 8 subjects that characterise procedural skills and thinking process to be subjugated by the following principles or guidelines--> Apply principles of safety, quality assurance and quality control in Clinical Microbiology Evaluate specimen acceptability. Describe morphology and physiology of microbes Identify and classify microorganisms Demonstrate sterile technique Perform and interpret antimicrobial susceptibility testing Select additional procedures based on preliminary results Correlate test results with patient condition(s) COURSE LITERATURE --> Typical Text: Textbook of Diagnostic Microbiology, 4th edition, Mahon C, Lehman D, Manuselis G Additional Texts/Readings: Laboratory Diagnosis of Infectious Diseases, Engelkirk P, Engelkirk,J Diagnostic Microbiology 12th edition, Forbes B, Sahm D, Weissfield A. Essentials of Diagnostic Microbiology, Shimeld, Lisa A. MATERIALS --> Standard Operating Procedure for Handling Laboratory Specimens (SOPFHLS) and so forth. All specimens will be handled using the "Standard precautions for blood and body fluids" recommended by public health (CDC) officials and government agencies. Practice will include: 1. Specimen brought into the MLT Laboratory will be carefully screened, in proper containers and labeled. 2. Proper protection will be used when processing specimen (lab coats, gloves and protective eye wear as appropriate). 3. Mechanical pipetting, use of safety engineering devices, and safe work practices at all times. 4. Decontamination of laboratory work surfaces at end of each exercise. Use bleach and/or phenol. 5. Decontamination or proper disposal of contaminated laboratory test materials. a. Blood and body fluids will be placed in biohazard bag in the trash. b. All needles, broken glass, and hemolets will be placed in a biohazardous sharps container and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. c. All gauze, Kim wipes and disposable materials contaminated with blood and body fluids will be discarded in a biohazard bag and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. 6. Decontamination of scientific equipment, i.e., electrodes, glassware, etc. 7. Hand washing after all laboratory procedures. 8. Accurate and proper recording and reporting of results. Reference: "Legal Implications of Universal Blood and Body Fluids Precautions", David L. Wing, Clinical Laboratory Science, Volume 1, No. 2, March/April 1988. " Needlestick Safety Act", OSHA Standards 2002 (or later version) 9. << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> 10. A standard academic laboratory manual accompanied by the likes of the following << https://www.mountsinai.on.ca/education/staff-professionals/microbiology >> 11. Microbial, Viral and pathogen software (check Goody Bag post) will follow applicable and practical mathematical modelling of cultures and so forth w.r.t. to median, nutrition and other environmental factors. All such along with lab studies of cultures --> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> 12. Lab attire and hygiene regulations In the laboratory, our first labs will address the basic use of the microscope, method of aseptic technique, how to perform basic staining procedures and how these common stains should appear under the microscope. This will familiarize students with several key stains and bacterial types. Students will also participate in the production of media to learn the associated math, contribute to the workload, and better understand the relationship between microbial growth and how media components that can enhance growth, select against, or differentiate different microbes Each student will perform a series of biochemical tests on an unknown bacterial sample to determine its identity. This demonstrates how microbes are characterized by their phenotypic characteristics and relates to how microbiology labs determine the causative agents in infectious disease clinical situations. Students will then work in pairs to develop a food microbiology hypothesis and design a guided experiment using serial dilution and plating methods to test their predictions with control and test food samples. To learn about normal and potentially pathogenic biota of the body, students will perform and analyse a series of experiments on their personal samples from five body sites. To study microbial control, students will test antiseptics, disinfectants and antibiotics for their effectiveness against common or personal microbial strains. Several experiments will also address the effectiveness of handwashing and alcohol-based hand sanitizing products. Mycology and parasitology will be introduced with prepared slides. Students will learn about the appearance and ratios of white blood cells by clinical differential white blood cell count methods on their own blood samples (or of prepared slides if preferred). There will be various types of microbes. Failure to observe safety rules as described above or as instructed will result in a lower grade and possible temporary or permanent expulsion from the laboratory. An unexcused lab without made-up work will result in 10% loss of laboratory points (40 points). An excused lab with the work made-up will result in a 5% loss of laboratory points. An excused lab without made-up work will also result in a 5% loss of laboratory points. Work may not be made-up without consultation with your instructor (you may not just simply come in on your own time unannounced or unplanned). Unknown Report = 60 points --> 1.You will be issued or able to select a clinically, food science, or environmentally relevant sample as an unknown. 2. You will perform staining and biochemical tests on the sample as a class. This will allow you to learn the necessary techniques and to generate data for your unknown sample. 3. A key of in-house lab test results for all possible unknowns will be provided and the creation of diagnostic keys and flow charts will be discussed. You will then devise a diagnostic key or flow chart for your unknown analysis. 4. The unknown report should consist of: a. Data chart: The chart of the results and interpretations of colony morphology, cell staining, biochemical, and oxygen requirements of the unknown. b. Flow chart: The chart should be followed by your diagnostic flow chart and the resulting determination of the unknown organism. c. Paragraph one: The analysis of the likelihood of the organism being correctly identified should then be discussed in paragraph format. This paragraph may also include mention of any difficulty in test results or interpretation analysis that could affect determination of the unknown. d. Paragraph two: Finally, you should look up the unknown in your lecture book or online. Discuss what clinical body site environmental site from which your unknown could have been isolated. If clinically important, discuss issues it could present for a patient. If an environmental or food microbe, discuss applications it may have. e. Resources: List any references used for the previous paragraph in a standard bibliography format. This reference list does not need to be on a separate page. 5. While exact identification is required for full credit, substantial partial credit will be awarded for correct interpretation of results and consistent analysis. Research Project = 40 points --> 1. This is a separate project from the Unknown Report. 2. These projects are to be designed, performed and analysed by pairs of students. The write-ups are to be done by each student independently. 3. The goal of this project is to test a hypothesis of personal interest. You and a partner will design a project to explore the issues of food poisoning and food intoxication and to practice hypotheses, predictions, sample dilutions, sample plating, scientific analysis and presentation of data. 4. Pairs of students are to create a hypothesis about the level of microorganisms in two different food samples that can be obtained or created from home. Next, predictions that can be tested should be made and a methods protocol drafted to be used in the lab set up. 5. The experiment should then be performed, and data collected. We will do this as a class together, but pairs of students will be working on different samples. Data should be analysed, and bacterial cells/ml or g of sample results determined for each concentration tested. Conclusions and further analysis of data should be considered. 6. The write-up for the research project should include an abstract and table of data collected. An entire paper is not necessary (and will result in a loss of points). An abstract is what scientists read to determine if they want/need to read a research paper any further. It is generally one page long only, but contains all of the key elements. It should contain the essential “nuts & bolts” of what was done, why it was done, how it was done, and what was found. Your abstract should be about 1 - 1.5 pages double-spaced in paragraph format and should address the following: a. What were you trying to test and why? b. What was your hypothesis? c. What were your predictions about the two different foods? d. How did you generally go about testing the food (hint, this should include the purpose of serial dilutions and not include a detailed about of every dilution you made)? e. Why do you want to calculate cells/ml or g from plates with ideally 25 – 250 colonies? f. What were your results (in cells/ml or g) and how did they compare to each other? g. What were your conclusions from your experiment? Did they support your hypothesis? h. What suggestions or modifications would you employ if you were to repeat the experiment? i. Will you change any food-related habits because of your results, or would you recommend behavioural changes to others? 7. A data table of all data collection should also be concluded. This explains how you generated final numbers for your abstract. 8. The complete assignment is due as listed on the laboratory schedule and should be typed up, printed out, and stapled to hand in. Figures can be done within the computer program used to create the document, or they may be taped into the document. Medical microbiology clinical culture lab reports = 50 points (10 points each) --> 1. These reports will be based on data and analysis on samples that you collect and process in lab. They should also provide insight into write-ups for clinical samples that might be analysed in a medical microbiology lab. They are designed to help you practice documentation of data and interpretation of results. 2. Lab reports to be filled out will be issued for the following systems: Bacteria of the Skin, Organisms of the Mouth, Bacteria of the Respiratory Tract, Bacteria of the Gastrointestinal Tract, and Bacteria of the Urogenital Tract. Academic well-being will be determined by the following --> Strictly enforced laboratory dress code Safe Laboratory behaviour and policies Laboratory safety devices Laboratory disposal Laboratory spills and accidents regulations and policies Academic Policies Laboratory attendance and participation policies Laboratory assignment policies Laboratory quiz and practical policies Prerequisites: Microbiology I & II Bacteriology: Did you know that without microorganisms, other life forms would not be possible? Did you know your body contains more bacterial cells than human cells? Did you know that one tablespoon of soil contains more microbes than people who have ever lived? Microorganisms are amazing! They were the first living cells on the planet and continue to be the most successful, living in every possible niche (including environments of pH 2 and temperatures over 100 degrees celcius) and utilizing every possible food source (even oil and plastic). During this course we will study the structure, physiology, and ecology of bacteria, viruses, protists, and fungi with an emphasis on bacteria. The course will specifically highlight areas of interaction between microbes and humans. We will gain experience with microbes in the laboratory as well. You will learn proper handling and identification techniques to help you complete longer term research projects. By the end of this course you will be able to: Remember basic taxonomic groups and key microorganisms in terrestrial, aquatic, and human environments. Connect microbe physiology to microbe ecology. Identify interactions between microbes and other living organisms. Culture, stain, identify and investigate microbes using aseptic techniques. Design a complete research project that incorporates both quantification and identification. Value the role of microbes in shaping the environment and human society. LABORATORY EQUIPMENT: Equipment: PPE-impervious lab coat, latex or nitrile gloves, closed toed leather shoes COURSE REQUIREMENTS: 1. Attend and complete all laboratory sessions. 2. Practice universal safety precautions during laboratory sessions. 3. Comply with program attendance code. 4. Complete assignments in a professional manner. 5. Complete all tests and exams given during the course in the allotted time and designated dates. COURSE GRADING: 3-4 Exams Lab Activity Lab Quizzes Lab Reports/Unknown Id/Skills Tests Lab Practicals There will be five (5) case studies presented during the course. They will presented in the “dropbox". Due dates will be assigned. Each case study has questions accompanying it that need to be answered correctly. Each case study will be worth 5 points. EXAMS --> There will be three mini-exams during the term. Each exam will be worth 40-50 points. The exams will consist of several short answer questions and one or two case studies to be analysed. LABORATORY-> We will spend the first few days learning essential skills for collecting, culturing, and analyzing bacteria from a variety of environments. You will complete two lab practicals to assess your knowledge of this information. The remainder of the semester will consist of three large projects: identifying three bacteria from a provided mix using traditional methods, identifying soil bacteria using DNA sequencing, and the completion of a research project of your own design. There will be opportunity to work with microscopic fungi and viruses, but the labs will emphasize bacterial cultures. Your attendance is expected at every class. You are responsible for any notes, announcements, or assignments that are missed. You are expected to read the assigned pages before class and be prepared to participate in class discussions. Late assignments will be accepted with a 10% penalty per class day late. PHASE 1 LABORATORY -Lab Policies; Microscope Use; Aseptic Technique; Cellular Morphology; Media Preparation; Simple Stain -Pure Culture Techniques; Colonial Morphology; Gram stain; Endospore Stain; Capsule Stain; Fungi and Protists -Oxygen Preference; Osmotic Pressure; Selective and Differential media; Antiseptics and Disinfectants; Disk Diffusion assay PHASE 2 LABORATORY -MViC; Sugar Fermentation; Sulfur Reduction; Oxidase; Catalase; Start Soil Project – plating dilution series -Freshwater microbes/Liquid Filtration; Food microbes/Standard Plate Count -Lab Practical I (7 days); Culture soil colonies on slants -Assign Bergey’s organism -DNA isolation and PCR from soil slants PHASE 3 LABORATORY -Bergey’s organism due; Receive unknowns/streak plate; Run gel of PCR results/Send for sequencing -Gram stain and slant unknowns; Continue research project -Inoculate unknowns on test media; ELISA test and epidemiology; Plaque assay; Kirby-Bauer antibiotic sensitivity test; Snyder assay; Hemolysin assay -Continue research project PHASE 4 LABORATORY -Lab practical II (7 days); Analyse DNA sequencing results -Continue research project; Finish unknown project; Finish soil microbe project -Soil project due LABORATORY QUIZZES (4) --> These quizzes will be small laboratory practicals where students will have to interpret test results that will be provided for them. Questions will be placed on index cards or printed sheet and the student will answer each question on an answer sheet. GRAM STAIN SKILLS TESTS (3) --> There will be 3 gram stain skill tests throughout the semester. The student will be graded on proper staining of the slide, interpretation and microscope focusing. ISOLATION SKILLS TESTS (3) --> There will be 3 isolation skill tests throughout the semester. The student will be graded on his/her ability to streak a colony on media using the proper technique to obtain isolated colonies. UNKNOWNS AND CLINICAL UNKNOWNS (2) --> May be adjusted based on organism availability and time. These unknowns will be simulated clinical samples in which the student has to correctly identify the organism using the correct battery of tests and submit the result on the unknown laboratory sheet provided. I. Introduction to Medical Bacteriology Review A. Classification of Medically Important Bacteria B. Steps Involved in Laboratory Diagnoses of Bacterial Infections C. Presumptive Vs Definitive Identification D. Laboratory Procedures Used in Diagnosing Bacterial Infections i. Culture Media ii. Gathering Information About an Organisms Phenotype MATERIALS --> Standard Operating Procedure for Handling Laboratory Specimens (SOPFHLS) and so forth. All specimens will be handled using the "Standard precautions for blood and body fluids" recommended by public health (CDC) officials and government agencies. Practice will include: 1. Specimen brought into the MLT Laboratory will be carefully screened, in proper containers and labeled. 2. Proper protection will be used when processing specimen (lab coats, gloves and protective eye wear as appropriate). 3. Mechanical pipetting, use of safety engineering devices, and safe work practices at all times. 4. Decontamination of laboratory work surfaces at end of each exercise. Use bleach and/or phenol. 5. Decontamination or proper disposal of contaminated laboratory test materials. a. Blood and body fluids will be placed in biohazard bag in the trash. b. All needles, broken glass, and hemolets will be placed in a biohazardous sharps container and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. c. All gauze, Kim wipes and disposable materials contaminated with blood and body fluids will be discarded in a biohazard bag and stored in the Prep Biohazard area for incineration and removal by commercial biohazard disposal company. 6. Decontamination of scientific equipment, i.e., electrodes, glassware, etc. 7. Hand washing after all laboratory procedures. 8. Accurate and proper recording and reporting of results. Reference: "Legal Implications of Universal Blood and Body Fluids Precautions", David L. Wing, Clinical Laboratory Science, Volume 1, No. 2, March/April 1988. " Needlestick Safety Act", OSHA Standards 2002 (or later version) 9. << www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi >> 10. << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> 11. << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> PERIODICALS --> 1. American Journal for Medical Technology 2. Laboratory Medicine 3. Medical Laboratory Observer (MLO) 4. Journal of Clinical Microbiology 5. Clinical Microbiology Reviews Typical test: TBA Typical Lab manuals (in unison) --> Designated lab manual + Bergey's Manual of Systematic Bacteriology COURSE OUTLINE: -Introduction/Cell Structure/Microbial Growth -Bacterial/Eukaryotic/Viral Taxonomy -Microbial Communication/Taxonomy Presentation Note: for bacterial communication the following articles may prove highly invaluable Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005; 21: 319 - 346 Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001; 55: 165 - 199 Taga ME, Bassler BL. Chemical communication among bacteria. Proc Natl Acad Sci U S A. 2003;100 Suppl 2 (Suppl 2): 14549 - 14554 Henke JM, Bassler BL. Bacterial social engagements. Trends Cell Biol. 2004; 14 (11): 648 - 656 -Bacterial Genetics -Bacterial Metabolism and Ecology -Microbes and Water Treatment -Microbes, Agriculture, and Food -Healthy Flora and Bacterial Infection -Epidemiology -Immune Responses/MMWR assignment due -Antibiotics and Resistance/Antivirals and Vaccines -Disease and Society -Disease Presentations -Disease and Society presentations Prerequisites: Microbiology II Virology: This class will introduce you to viruses. We will emphasize the development and life-cycle of all types of viruses, but will cover some aspects of epidemiology. We will especially emphasize (lambda), Hepatitis C (HCV), Feline Leukemia Virus (FeLV), Influenza, HIV, MERS and SARS in the course. Typical text --> Introduction to Modern Virology, 6th Edition N.J. Dimmock, A.J. Easton, and K.N. Leppard Blackwell Scientific Publications, 2007 Journal Articles --> May have to incorporate journal articles as well Software --> Microbial, Viral and pathogen software (check Goody Bag post). Many lectures AND labs will incorporate software --> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> Organic Chemistry, Biochemistry & Molecular Biology software --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Grading --> - The Labs will count for 55% of the final grade. You will need to write up the results for each experiment. A separate handout explains the lab writeups. Attendance at all the labs is required, with points deducted for missing them. The lab write-ups are due on Friday’s, one week after the lab has finished. - Then 45% of the grade will come from your scores on 3 midterms. They cover the lecture and (most, not all) lab material. Course Outline --> WEEK 1 Introduction. Growth and assay of viruses WEEK 2 Methods of studying viruses. Methods of identification of biochemical molecules in viral structure, cell interactions, infiltration and (high-jacking) replication. Structure of viruses WEEK 3 Classifying viruses. Lab-Collect soil phages. Wear sensible shoes. Attachment and penetration WEEK 4 Replication of DNA virus genomes WEEK 5 Replication of Retroviral genomes Gene expression in DNA viruses WEEK 6 Review for Midterm #1 Gene expression in DNA viruses WEEK 7 - 8 Gene expression in DNA viruses. Gene expression in RNA viruses. Assembly of viruses WEEK 9 Assembly of viruses. The immune system. Virus-cell interactions WEEK 10 Virus-host interactions. Lambda WEEK 11 Virus latency. Transmission of viruses WEEK 12 Review for Midterm #2 Transmission of viruses. Viral evolution WEEK 13 Viruses and Human Diseases WEEK 14 Carcinogenesis and tumor viruses. Vaccines and antivirals WEEK 15 (viral communication and virus-virus interactions) Erez, Z., Steinberger-Levy, I., Shamir, M. et al. Communication Between Viruses Guides Lysis–Lysogeny Decisions. Nature 541, 488–493 (2017) Note: the prior article may just be one conveyance, where possibly other articles or texts give further elaboration. Seek out texts and journal articles for analysis, modelling and biochemical process. WEEK 16 Vaccines (viral vector, plasmid DNA, mRNA and recombinant nanoparticle) and antivirals Strategy and mechanisms (can make use of mentioned software for intimate investigation) Prions WEEK 17 Review for Midterm #3 LAB SCHEDULE --> In the labs, you will a perform research project. In one, we will purify and characterize phages from whatever environments. The emphasis in the labs is not on the correct results, but instead, it is on understanding what you are doing and why you are doing it. By understanding what the experiments can and cannot do, you can then understand original papers and seminars in molecular biology and related fields. You also get some feeling for what types of problems can occur and be solved in labs. Since exact results in the labs are not important, do not alter your data to make it look better. Preparation for the labs is important. Reading the labs over before class will not only reduce mistakes in the lab, but it will also help you work more quickly. In general, try to understand the types of uses for each technique, the range of sensitivity for each technique, and the advantages and disadvantages of similar techniques. Try to think of other uses of the experiments and techniques. We only have enough time to use a technique one way, but most of them can be used for many types of experiments. Note: construct isolated sealable tents, where professional safety and hazard protocols are present and in place, if need be. Note: video recording of labs is permissible, granted that the given regulations and protocols are upheld. Lab Week 0 Safety and walkthroughs Applicable and practical mathematical modelling of cultures w.r.t. to median, nutrition and other environmental factors; accompanied with modelling software. NOTE: such may also be involved with other labs besides lab week 4, say, lab week 7 - 12. Lab Week 1 T4 plaque assay Lab Week 2 Isolation of soil phages Lab Week 3 Soil phage. Isolation of soil phages Lab Week 4 Growth of phage Augmented by review of some modelling skils from Lab week 0 accompanied with modelling software Lab Week 5 Isolation of phage nucleic acid Lab Week 6 Analysis of phage nucleic acid Lab Weeks 7 – 9 Further analysis of phage Lab Weeks 10 – 11 Detecting viral sequences (DNA and RNA) Lab Weeks 12 – 13 Role of proteins PART A Proteins and host cell membranes Protein identification by mass spectroscopy or Edman Degradation. Protein-protein, protein-nucleic acid, protein-RNA and protein-lipid interactions determine: (1) structure of virus particles (2) the synthesis & expression of virus genomes and (3) the effects of viruses on the host cell. Relevant to the mentioned 4 types of interactions what methods and experiments discovered/confirmed the mentioned interactions towards (1) through (3)? Target sites, functional groups, protein dynamics w.r.t counterpart concerning the four types of interactions. Example viruses of interests: common cold viruses, Influenza viruses, HBV, HCV, Feline Leukemia Virus (FeLV), measles, Ebola, Zika, HIV, MERS, SARS, etc., etc. Then to make use of software out of the following for building visualizations and simulations: << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Highlighting of the following: Classification Symptoms and hazards Anatomy Genomes Sequences transferred to replicate Infiltration process (biochemical processes) Nucleous engagement and tasks Disease ailments (and pathway descriptions leading to ailments) Successful counter-responses against cellular penetration Antibodies tactics Vaccine guide development and testing Interesting article for measles: Mina, M. J. et al. (2019). Measles Virus infection Diminishes Pre-existing Antibodies that Offer Protection from other Pathogens. Science 01 Vol. 366, Issue 6465, pp. 599 - 606 Lab Week 14 PART A: Subtyping (lab activity with 1 or 2 methods) Arens, M. (1999). Methods for Subtyping and Molecular Comparison of Human Viral Genomes. Clinical Microbiology Reviews, 12 (4) 612-626 PART B: Software for genomics development following: RStudio + R packages for genome data development (BiocManager, ChemmineR, dPCP, MAPITR) --> Thodberg, M., & Sandelin, A. (2019). A Step-by-Step Guide to Analysing CAGE Data using R/Bioconductor. F1000Research, 8, 886 Cao, Y., Charisi, A. et al. (2008). ChemmineR: A Compound Mining Framework for R. Bioinformatics (Oxford, England), 24(15), 1733–1734 < manuals.bioinformatics.ucr.edu/home/R_BioCondManual > < cran.r-project.org/web/packages/BiocManager/vignettes/BiocManager.html > Students may be given anonymous pathogen sequences. Interests to be out of the most predominant or advanced or accurate methods recognised today. Lab Week 15 Vaccines and pathogen evolution (lab activity with 1 or 2 methods for strain recognition/differentiation) PART A (overview): Ojosnegros, S. & Beerenwinkel, N. (2010). Models of RNA Virus Evolution and their Roles in Vaccine Design. Immunome Research, 6 (Suppl 2), S5 PART B (overview): Vaccines and pathogen evolution Sanjuán, R., Domingo-Calap, P. (2016). Mechanisms of Viral Mutation, Cell. Mol. Life Sci. 73, 4433–4448 Hanley K. A. (2011). The Double-Edged Sword: How Evolution Can Make or Break a Live-Attenuated Virus Vaccine. Evolution, 4(4), 635–643 PART C (overview) Viral Mutation Rates Will have lab investigation of 2 - 3 “vanilla” viruses to determine mutation rates based on the following article: Sanjuán, R., Nebot, M. R., Chirico, N., Mansky, L. M., & Belshaw, R. (2010). Viral Mutation Rates. Journal of virology, 84(19), 9733–9748 PART D (activity) Database viral mutation research to develop. Replicate then pursue other viruses: Yun-Xin Fu, (2001). Estimating Mutation Rate and Generation Time from Longitudinal Samples of DNA Sequences, Molecular Biology and Evolution, Volume 18, Issue 4, Pages 620–626 General Approach Data Collection: Gather longitudinal samples of DNA sequences from the population of interest. These samples should span multiple time points to capture the changes in the genetic makeup of the population over time. Sequence Alignment Phylogenetic Analysis Molecular Clock Analysis Generation Time Estimation Statistical Methods: to quantify the uncertainty associated with mutation rate and generation time estimates. This may involve Bayesian methods (or bootstrapping) to generate confidence intervals. Model Selection: Choose appropriate evolutionary models and clock models based on the characteristics of the studied population. Different viruses or organisms may have different mutation rate patterns, and selecting an appropriate model is crucial for accurate estimation. Quality Control: Implement quality control measures to ensure the reliability of the results. This includes checking for sequence errors, contamination, and other potential sources of bias. Validation: Validate the estimates by comparing them with independent datasets or using alternative methods. This helps assess the robustness of the results. Prerequisites: Microbiology II, Cell Biology, Organic Chemistry I It will be assumed that students are well on their way passes Biochemistry I and are committed to their advancement obligations. Tissue Culture & Virology Lab: This course is different to the Virology course in the sense that this course involves highly thorough protocols and operations in active labs with cell and viral cultures. The class is divided into three modules: cell culture, fundamental virology, and identification and characterization of an unknown virus sample. Module I --> Cell culture. The objective of this module is to learn fundamental cell culture techniques including sterile technique, cell maintenance, cell-splitting, and freezing cells. Module II --> Fundamental virology. The objective of this module is to learn standard assays used in virology studies such as virus amplification, plaque assays, host-range studies, and ELISAs. In addition to learning the technical details, we will focus on how to interpret, analyse, and evaluate experimental results. Module III --> Identification and characterization of unknown viruses. The objective of this module is to identify and characterize an unknown virus sample based on the techniques and analysis methods learned in the first half of the semester. You will summarize this module by writing a journal article that emphasizes the significance and results from your work. Experiment worksheets --> Each experiment will have an accompanying worksheet. These worksheets are meant to be filled in as you are performing the experiment and guide you as you perform the experiment. These worksheets also provide a place to record data, do calculations, and analyse your results. During Module III the worksheets will guide you in identifying your unknown virus. Please see the syllabus and calendar for when worksheets are due. Homework --> The homework assignments are a combination of concepts and principles, data analysis, and are based on the lab and pre-lab lectures (see the syllabus and calendar for when worksheets are due). The homework assignments are meant to be a check-point to ensure you understand the main concepts of what is being taught. Exams --> Two exams are given in this course. The first exam is before spring break. The exam is short answer and covers the principles and concepts taught during Modules I and II. The second exam is at the end of the semester and is a lab practical. Here you are shown actual results and asked to interpret, analyse, and draw conclusions. Lab worksheets, pre-lab lectures, and homework assignments are your best guide for preparing for these exams.If you have university documentation showing you need extra time for an exam, please let me know within the first two weeks of class. Unknown virus write-up --> For Module III, Identification and Characterization of an Unknown virus, you will summarize your results as a scientific paper. You will detail how you determined the identity of your unknown virus and its properties. Results from each experiment will be summarized as a figure or table, and an appropriate introduction and discussion should be included. Guidelines for how to write this paper are at the end of the lab manual and we will discuss this more during the second half of the semester. To help you learn about scientific writing, you will be required to turn in at least one “Results” section and its corresponding Materials and Methods” section before turning in the final unknown lab write-up. Lab performance --> There are many times during the semester that you will need to come in during non-lab hours to finish an experiment or record results. Experiments can be complicated, require reagent preparation beforehand, and require care since you will be working with live virus. Your lab performance grade will be based on ability to maintain cell lines, preparing for an experiment, and returning to finish an experiment. This will be determined by both your AI and me. The five different components contribute to your final grade --> Experiment worksheets 500 points (25 points/each, 20 in total) Homeworks 400 points (50 points/each, 8 in total) Exams 400 points (200 points/each, 2 in total) Unknown virus write-up 100 points Laboratory experiments --> An experiment may take 1-2 weeks to finish. Many labs require a few days of preparation (usually getting your cells ready), a day to do the experiment, and another day to obtain and analyse the results. To help you keep track of the experiments a detailed schedule has been made. This is in the lab manual and on “Oncourse” as a .pdf file. The schedule is only a guide and should be used as the first step in planning your experiments. It is your responsibility to have your cells ready on the day needed and to come into lab and record your results when necessary. Students will work in groups of two. Each group will work in their own hood and be responsible for cleaning the hood, discarding trash and bringing pipettes and biohazard waste to front of the room at the end of each lab period. Each group will receive some initial supplies the first day which they are responsible for maintaining. Both partners must attend class and participate in the lab. If you have to miss a class (illness, job interview), please let your lab partner and myself know as soon as possible. Typical texts --> Tissue Culture and Virology laboratory, by Tuli Mukhopadhyay, published by RLSimonson Studios Gnaguly, S. (2014). A Laboratory Manual on Virology and Tissue culture Techniques. Narendra Publishing House Supporting text: O’Kelly E. (1998) Cell Culture and Diagnostic Virology. In: Clynes M. (eds) Animal Cell Culture Techniques. Springer Lab Manual. Springer, Berlin, Heidelberg Journal Articles --> May have to incorporate journal articles as well Software --> Microbial, Viral and pathogen software (check Goody Bag post) will follow applicable and practical mathematical modelling of cultures and so forth w.r.t. to median, nutrition and other environmental factors. All such along with lab studies of cultures << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> Course Outline --> Week 1: Experiment #1: Introduction to cells: importance of sterile technique. Check in & get supplies. Discuss lab safety and biosafety. Informational sheet. Continue/Finish Experiment #1. Experiment #2: Introduction to cells: visualizing different cell types and determining cell viability and concentration. Worksheet Experiment #2 due Week 2: Finish Experiment #1. Experiment #3: Introduction to cells: learning to passage and maintain cells. Continue Experiment #3. Worksheet Experiment #1 due. Week 3: Continue Experiment #3. Experiment #4: Introduction to cells: freezing and storing cells. Worksheet Experiment #3 due. Week 4: Continue Experiment #4. Experiment #5: Introduction to viruses - determining infectivity by plaque assay. Finish Experiment #4. Finish Experiment #5 Worksheet Experiment #4 due Worksheet Experiment #5 due Week 5: Experiment #6: Introduction to viruses - propagating virus samples Continue Experiment #6 Finish Experiment #6 Week 6: Experiment #7: Introduction to viruses - determining how MOI influences infectivity Experiment #8: Introduction to viruses - determining which hosts are susceptible to viral infection Worksheet Experiment #6 due Continue Experiment #7 and #8 Finish Experiment #7 Finish Experiment #8 Experiment #9: Introduction to viruses - determining viral growth kinetics Worksheet Experiment #7 due Worksheet Experiment #8 due Continue Experiment #9 Week 7: Continue Experiment #9 Finish Experiment #9 Worksheet Experiment #9 due Week 8: Protein-protein, protein-nucleic acid, protein- RNA and protein-lipid interactions determine the structure of virus particles, the synthesis and expression of virus genomes and the effects of viruses on the host cell. Target sites, functional groups, etc. etc. Can make use of software out of the following: << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Experiment #10: Characterization of viruses - ELISA assay Worksheet Experiment #10 due Experiment #11: Characterization of viruses - pH and lipid sensitivity Experiment #12: Characterization of viruses - hemagglutination assay Worksheet Experiment #11 due Finish Experiment #11 Finish Experiment #12 Week 9: Experiment #13: Characterization of viruses - electron microscopy Worksheet Experiment #11 due Worksheet Experiment #12 due Worksheet Experiment #13 due Week 10: Experiment #14: Identification of unknown virus Determine host-range of unknown virus Follow up: As needed for all experiments during rest of semester Turn in worksheets as you finish experiments Week 11: Characterization of pH and lipid sensitivity of unknown virus Start amplifying unknown virus Week 12: Determination of nucleic acid type in unknown virus Week 13: Determine infectivity of amplified unknown virus by plaque assay Methods and results for at least one experiment Week 14: Repeat experiments if necessary Last day for experiments Clean-up Check-out Last day to turn in worksheets #14 - #20 Week 15 EXAM II, Lab practical Write-up for Experiment #14 due Co-requisite or Prerequisite: Virology Prerequisites: Microbiology I & II, Cell Biology, Organic Chemistry I Microbiology of the Digestive System: Course Description: Study of microorganisms of the rumen and hind-gut of mammals, and their contributions to nutrient utilization of the host animal. Other topics will include nutrient metabolism by microbes, species interactions, and techniques for identification of species and populations of microbes. Student Learning Outcomes --> Develop a solid understanding of the microorganisms of the digestive tract of mammals Be able to describe how microorganisms contribute to nutrient utilization of mammals Be able to identify common digestive tract microorganisms Describe how species of microorganisms interact with one another Describe techniques to identify species of microorganisms Microbial growth models with environmental parameters versus observation (lecturing and labs) Texts--> Will make use of various professional sources that are highly comprehensive or extensive in the course topics. References --> Verhoeckx K. et al. (eds) The Impact of Food Bioactives on Health. Springer, Cham (2015) Hillman, E. T., Lu, H., Yao, T., & Nakatsu, C. H. (2017). Microbial Ecology Along the Gastrointestinal Tract. Microbes and Environments, 32(4), 300–313 Karasov, W. H., & Douglas, A. E. (2013). Comparative Digestive Physiology. Comprehensive Physiology, 3(2), 741–783 Labs --> One isn’t interested in elementary or junior high experiments; will be left to students to look up such. Nevertheless, the following example articles make strong guides for lab experiments (make appropriate order and w.r.t. course topics succession): --Processing of Vitamins, Minerals, Proteins, Sugars, Starches and Fats. Environments for stability and solubility with verification through biochemistry lab activities. Includes what parts of the digestive tracts are responsible for processing; consideration for chemical reactions and by-products as well. Temperature, pH and other variables may be considered as well. If microbes are highly prevalent in process, must have detailed analysis, modelling and simulation of the biochemical processes. NOTE: software encountered in Biochemistry I will be applied in this lab. --Cheng, C. H. K. Laboratory Experiments on the Actions of Digestive Enzymes, Biochemical Education 20(1) 1992 --Robic S. (2010). Laboratory Exploration of Survival of Probiotic Cultures Inside the Human Digestive Tract Using in vitro Models. Journal of Microbiology & biology education, 11(1), 50–55 --Liu L, Firrman J, Tanes C, Bittinger K, Thomas-Gahring A, Wu GD, et al. (2018) Establishing a Mucosal Gut Microbial Community in vitro Using an Artificial Simulator. PLoS ONE 13(7): e0197692. --Brodkorb, A., Egger, L., Alminger, M. et al. INFOGEST Static in vitro Simulation of Gastrointestinal Food Digestion. Nat Protoc 14, 991–1014 (2019). --Minekus M. (2015) The TNO Gastro-Intestinal Model (TIM). In: Verhoeckx K. et al. (eds) The Impact of Food Bioactives on Health. Springer, Cham --Bergmann, M. and Fruton, J. S. Some Synthetic and Hydrolytic Experiments with Chymotrypsin. J. Biol. Chem. 1938, 124: 321-329 --Other labs will involve particular microbial organisms involving “specified nutrition” and environmental parameters (temperature, pH, etc. etc. etc.) with dependent variables such as growth rate, rate of processing, etc. etc. etc. NOTE: software mentioned in Environmental Microbiology and Clinical Microbiology may be applicable in this lab. Grading: 3 Exams: 60% Labs: 40% Course Topics: I. Introduction to Microbiology II. Bacteria of the Digestive System III. Protozoa of the Digestive System IV. Fungi of the Digestive System V. Interactions of Digestive System Microorganisms VI. Microbial Techniques VII. Carbohydrate Fermenting Microorganisms VIII. Microorganisms Involved in Nitrogen Metabolism IX. Microorganisms Involved in Lipid Metabolism X. Methanogenic and Acetogenic Bacteria Prerequisite: Biochemistry, Microbiology II, ODE. Note: try not to take this course at the same time with Clinical Microbiology and Environmental Microbiology. Tissue Engineering Course is different from the Tissue Culture & Virology course. This course DOES NOT have virology applications. A standard tissue engineering course can be developed with not much trouble. However, many are not innovative enough to recognise that undergraduate students with laboratory skills from prior courses are highly capable of successfully completing tissue engineering laboratory activities. The skills students have serve to engage or immerse themselves in economic sensibility (professional opportunities, recognising the high value in their skills and self-worth, etc., etc.). Apart form standard lectures for crucial education with applications in medicine and engineering, interest and access can be amplified if students’ skills are put to use. From the given articles below one can recognise that the materials and tools required for tissue engineering labs are often standard and relatively inexpensive equipment used in other biological sciences courses. Department to build a course tailored to incorporating activities observed in such articles --> Saterbak, A. (2002). Laboratory Courses Focused on Tissue Engineering Applications. Proceedings of the 2002 American Society for Engineering Education Annual Conference & Exposition Copyright © 2002, American Society for Engineering Education Bodnar, C. et al (2018). Implementation and Assessment of an Undergraduate Tissue Engineering Laboratory Course. Education for Chemical Engineers 24, 52 – 59 There’s also the evolving science and technology of lab produced meat. Some of the example articles following provide strong descriptions of experiments that can be implemented in course --> Datar, I. and Betti, M. (2010). Possibilities for an In-Vitro Meat Production System, Innovative Food Science & Emerging Technologies. 11 (1): 13–22. Adamski, M. et al (2018). Two Methods for Decellularization of Plant Tissues for Tissue Engineering Applications. Journal of visualized experiments: JoVE, (135), 57586. Ben-Arye, T. et al. (2020). Textured Soy Protein Scaffolds Enable the Generation of Three-Dimensional Bovine Skeletal Muscle Tissue for Cell-Based Meat. Nat Food 1, 210–220 Kang, D. H. et al (2021). Engineered Whole Cut Meat-Like Tissue by the Assembly of Cell Fibers Using Tendon-Gel Integrated Bioprinting. Nature communications, 12(1), 5059. NOTE: one isn’t restricted to any particular activities in the articles. Based on analysis and experience one can introduce other experiment assignments for robustness. Grading Homework Quizzes Labs 3 Exams Prerequisites: Cell Biology, Biochemistry I, Genetic Engineering & Technology, Molecular Biology I
B. METABOLIC BIOLOGY (BIOCHEMISTRY) Students will proceed with courses based on prerequisites they have successfully completed with satisfactory grade requirement. Concentration Curriculum: --Core Courses Scientific Writing I & II, General Biology I & II, General Chemistry I & II, Organic Chemistry I & II (check chem post), Biochemistry with labs, Biostatistics I & II, Advanced Statistical Modelling and Machine Learning for Biostatistics --Professional Necessities Food Chemistry, Organic Synthesis Laboratory, Cell Biology (with labs), Genetic Engineering & Technology, Comparative Genomics, Molecular Biology I & II, Metabolism, Molecular Control of Metabolism & Metabolic Disease, Bioprocess Engineering, Bioprocess Engineering Lab, Drug Metabolism, Protein Engineering, Metabolic Biology Research. --Mandatory Courses Calculus for the Biological Sciences I & II, ODE, General Physics I Some course descriptions: Biostatistics I Course concerns probability and statistics applied to problems in biology, industrial/occupational health, and epidemiology. Use of statistical software R for data analysis is emphasized extensively. Note: this course is designed for students majoring in the biological sciences with a second term calculus background. Through the extensive use of practical examples, this course is expected to motivate and teach students statistics knowledge that would be helpful for their major study. The computer program R is the standard statistical program for this course. Students will use R to complete data analysis projects. R can be downloaded and installed on your personal computer for free following instructions at http://www.r-project.org/. In addition, the R environment will be augmented by RStudio interface with other R packages. This course covers fundamental concepts in probability and statistics, including data description, design of experiment, probability rules and distributions, statistical inference and linear regression. Definitions will be learned through real-world examples and applications. Besides these traditional materials and subjects, topics and methods that are particularly applicable to the biological sciences will be introduced. Again, much focus on the applications of statistical ideas to realistic data and practices. Students are expected to use materials learned from this course to guide statistical practice for their major studies in the future. After successfully completing this course the expectation is that students will be able to: 1. To grasp concepts in probability and be able to apply basic probability rules, distributions, and laws to solve conceptual statistics questions 2. Use statistical guidelines and common sense to interpret the process of data collection, description and analysis, and to design statistically sound experiments 3. Learn various statistical inference techniques and be able to select appropriate methods for specific data sets and scientific purpose 4. Link the course materials with real-life examples, and explore the opportunities for other biological applications 5. Interpret statistical reports and carrying out data analysis using R. Several data projects will be assigned during the semester. Independent work is expected. This is not a course of “pen and paper finesse” succeeding the composition of gunk and bamboozle on a writing board. One can’t be successful in statistics by only writing down theory. Practice with an environment that applies intelligence and engagement is essential. There’s no point in doing statistics if one doesn’t know how to acquire and manipulate real data. Real data realistically outnumbers the fingers and toes one possesses. Most of grading will be based on projects (having commentary descriptions) accompanied by the analytical process development description done in a word processor. Typical Texts --> Will make use of R language Statistics texts under CRC Press and Springer publishing Tools --> R language and R Studio Note: a calculator at times may prove useful for the idealistic or the “synthetic” customary questions. “R Monograph Notebook” --> Students should maintain a notebook as they proceed through the course and learn how to do analyses in R. This assignment involves a notebook that lists the syntax and provides a brief explanation of each function that students learn during the course. Instructor will assign R maturity development questions to be in tune with course progress; you will only be allowed to use your monograph to assist with assigned questions. The notebook will be handed in near the end of the semester and handed back to the students after grading. Such a notebook can be an extremely useful resource both during and after the course to quickly refresh one’s memory on the details of a particular function. Design with R most likely will vary among students. Poor development in a such a notebook may or may not correlate with poor grades. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS. Grading --> Problem Sets 20% R Labs 25% R Activities 0.7 Will involve all course topics R Monograph Lab Notebook 0.3 R maturity development questions End of term lecturer observation 3 Exams 30% Assigned Projects 25% Exams --> Limited open notes. I don’t like setting up myself and students for embarrassment; you are not perfect with statistics, so expect exams to be primarily knowledge based and the calculus related fodder. Most of your development will come from homework and labs; it is what it is. Note: limited open notes. Students may be more comfortable with certain R packages. Again --> Several data projects will be assigned during the semester. Independent work is expected. Course Outline --> Introduction to statistics, data and R Statistical Measures and Summary Statistics for data sets Data acquisition: Sources/databases (ecological & biological), file types, APIs, etc. Data Wrangling Summary Statistics Applied probability theory Axioms of probability Modelling frequencies and establishing densities Simulating random variables from real experiments Probability distributions and properties Law of Large Numbers and the Central Limit Theorem Introduce the Law of Large Numbers (LLN) Central Limit Theorem (CLT). Identify Exponential, Poisson and Binomial data and respectively determine in a manner to confirm LLN and CLT. Routledge, R., Chebyshev’s Inequality, Encyclopaedia Britannica Is there too much reliance on assuming normal or Gaussian distribution? Towards Chebyshev’s inequality what amount of repetition (regarding LLN and CLT) of an experiment is adequate towards Chebyshev becoming relevant? Overview and goals of various concentration inequalities (just a survey). Sample Estimates Chi-square distribution The bottom line is to establish the flow of the uses competently with applications involving real raw data. Comprehending categorical data sets Organisation of data and sensitivity of categories concerning traits of interest. Test for independence McHugh ML. (2013). The chi-square Test of Independence. Biochem Med (Zagreb). 23(2): 143-9. Test of homogeneity Test of variance Applications of the Chi-Square distribution with confidence intervals T-distribution Kim T.K. & Park J. H. (2019). More About the Basic Assumptions of T-test: Normality and Sample Size. Korean J Anesthesiol. 72(4): 331-335. Sample size determination Population parameter estimation Confidence intervals Directly logistical to understand what you’re doing in R F-distribution Assumptions for the F-distribution Relevance to the biological sciences (active data immersion) Note: not textbook finesse, rather how and when actively. Goodness of fit: fit of distributions Summary Statistics Skew and Kurtosis P-P and Q-Q Statistical Tests Definition, Null hypothesis One-sided & two-sided tests of hypothesis Types of test statistics Comprehending critical values for ideal distributions Significance levels Critical values for real raw data sets Does your data exonerate ideal distributions? Chi-Square Test Kolmogorov-Smirnov Test Anderson-Darling test Shapiro-Wilk Test MLE and Method of Moments Manual tasks will be limited to at most 4 element data sets Computational logistics for large data sets followed by implementation Review/probe data for goodness of fit module for appropriate distribution You may be tasked with distribution determination before parameter/point estimation Hypothesis testing (exploratory, sans assumption of distribution) Note: as aspiring biologists I can’t give “zombie textbook problems” and expect you to relate to a profession tangibly and fluidly. You will be exposed to raw professional data from various sources. You will develop the four mentioned steps. You should ask yourselves if the hypotheses are practical as well. Why is normal distribution assumed? Will be exploratory rather than zombie problems. Namely, knowledge and skills from Goodness of fit module. Then proceed with the following: 1.State the two hypotheses so that only one can be right 2.Formulate an analysis plan, outlining how data will be evaluated 3.Carry out the plan and physically analyze the sample data 4.Analyse the results and either reject the null, or state that the null is plausible, given the data 5.Directly logistical to understand what you’re doing in R Test of Proportions (exploratory) Differences in population (mean and median) Comparisons of variances Confidence limits for means. Does it require normality? Correlation (includes misuse of types and resolutions) Pearson Correlation Crucial Conditions Structure Implementation Spearman Correlation Crucial Conditions Structure Implementation Kendall Correlation Generating heat maps. The ggpairs() function Bivariate Regression Multiple Regression Model components Methods to select variables OLS MUST: Summary Statistics and forecasting Analysis of Covariance Must be exploratory, else it’s toxic Non-parametric statistics Resampling methods Falsified Data Hartgerink C, Wicherts J, van Assen M (2016). The Value of Statistical Tools to Detect Data Fabrication. Research Ideas and Outcomes 2: e8860. Al-Marzouki, S., Evans, S., Marshall, T., & Roberts, I. (2005). Are these data real? Statistical methods for the detection of data fabrication in clinical trials. BMJ, 331(7511), 267–143. Yamamoto, K., & Lennon, M. L. (2018). Understanding and Detecting Data Fabrication in Large-Scale Assessments. Quality Assurance in Education, 26(2), 196–212. Prerequisites: General Biology II, Calculus II Biostatistics II --This succeeding course in the sequence will have more emphasis on incorporating journal articles and real world experiments. --Students will have to orchestrate inquisitions by exploratory data analysis and statistical methods involving R. There will be assigned data sets and journal articles to do just that. --This is not a “pen and paper course”. Texts and journal articles will cater to subjects both from prerequisite and this course. Means of data retrieval and manipulation are crucial; it may be the case that the data desired in inaccessible, hence students will have to resort to alternative data sources that yields much different conclusions. NOTE: personally refresh your knowledge and acquired R skills from calculus and Biostatistics I alongside ordained reacquaintances in course. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS, AND KEEP YOUR DAMN BUSINESS . Assessment --> Assignment Sets (prerequisite & current level from multiple sources) 15% Analytical and R based 3 Exams (prerequisite & current level) 30% Labs + Data Analysis Term Project 40% 2 Field Inquisitions with R 15% Conducted Journal Articles Computational Inquisitions Supporting data sets to be provided Gov’t administration field experiments inquisitions Assignment Sets --> Will be reacquainted with prerequisite tasks, prerequisite projects AND current level tasks (analytical and R computational). Exams --> Exams will have the same manner of administration and activities as exams from prerequisites. Yet, consisting of both prerequisite tasks AND current level tasks. Limited open notes. LABS WITH R --> Data Processing Data Assimilation (ecological & biological), file types, APIs, etc. Data Wrangling Descriptive Statistics Hypothesis Testing (advance practice from prerequisite) Expt. Design, Multiple Comparisons Regression (Mult. Reg. and Dummy) ANCOVA MANCOVA Non-parametric Statistics (advance practice from prerequisite) Clustering PCA and Kernel PCA Comparing & Averaging Models Analysis of Trait Evolution Fitting models of Trait Evolution TERM PROJECT --> The term project has been broken down into multiple components due throughout the semester to provide further guidance for students. On given date, students will select a dataset to use for their term project. Students can either provide their own dataset (if they have collected data during their research), or will be given the opportunity to analyse a complex dataset supplied by faculty as their term project. For the Hypothesis Activity (given due data), students will take a close look at their dataset and formulate biological hypotheses that they would like to test statistically. The assignment will be handing in these hypotheses. For the Experimental Design assignment (given due), students will outline which analyses they will use to test their biological hypotheses and provide the specific explicit statistical hypotheses that they will test. The Term Project Report (given due having additional 1 week collection buffer) will be written after students complete their analyses. The report will include a Statistical Methods and a Results section, complete with tables and figures. Methods should include sufficient detail to redo the analyses. The results should include everything necessary for interpretation of their analyses and data, but not superfluous material. Term Project Reports for all students should include a title page with a title, student name, course number and name, and assignment name. The text of the report should be double spaced, with indented paragraphs, 1” margins, 12pt Times New Roman Font, and page numbers. Tables should be single spaced with headings above each table. Figures should have captions below each figure. Figures and tables can be embedded in the text or provided at the end of the document. Literature cited should follow the format for the journal Evolution. Assignments that do not follow these formatting instructions will be returned to the student for correction prior to grading. NOTE: most labs done serve as structure for your HA and ED NOTE: I will be collecting your R development (having sensible commentary) for the term project in PDF along with the term report in PDF. Finally, students will give a short, in-class presentation about their study, analyses and findings. Presentations will be in PowerPoint. MAJOR COURSE TOPICS --> Methods of data acquisition, data wrangling and summary statistics (prerequisite reinforcement) Goodness of Fit (prerequisite reinforcement) MLE and MoM Hypothesis Testing (prerequisite reinforcement) Experimental Design & Sampling Regression (multivariate) OLS review (variable selection, summary statistics, forecasting) Quantile Regression compared to Least Squares regression Variable selection, summary statistics, forecasting ANCOVA MANCOVA Non-parametric Statistics Resampling Techniques Clustering (K-means or DBSCAN?) Principal Component Analysis (PCA) and Kernel PCA Model Selection & Likelihood (emphasis on computational logistics and implementation) Phylogenetic Regression Extensions of Phylogenetic Statistics Prerequisite: Biostatistics I Advanced Statistical Modelling and Machine Learning for Biostatistics This course explores advanced statistical modeling techniques and machine learning methods as applied to biostatistical problems. Topics include generalized linear models, hierarchical modeling, Bayesian statistics, and the integration of machine learning algorithms for analyzing complex biological and health data. Note: 2 lectures per week, with approximately 2 hours per lecture. Assignments -- Assignments will be quite laborious in the interest of sustainability with knowledge and skills through your journey in biostatistics. Each assignment will comprise of the following elements: A. problems and tasks encountered in both Biostatistics I & II. Such problems and tasks will also make extensive use of R. As well, being advanced biostatistics students, projects from Biostatistics I & II can/will also be considered basic assignments as well. B. Course level assignments to such given course topics. Good emphasis on ability to comprehend and specify the transition from prerequisite skills/tools to course level tools/method/skills; then implementation. Will also make extensive use of R. Data Science Basics Quizzes -- For the Data Science Basics module there will be handwritten quizzes to test knowledge, comprehension, appropriateness and T/F. Exams -- Exams will account for all modules. Assignments will be strong foresight of what’s to appear on exams. You will be making extensive use of R with open notes for all course modules. Exams will feel like projects where each “project” will involve multiple modules. Make-up Student Project -- Applying Advanced Models to Biostatistical Data. Concerns students who are interested in making up lost weight towards their final grade; the better you did in this course, the lower the value. Can regain up to 5% for final grade. Students will be given a sack to randomly (and blindly) pick a project. Students will have until 2 days before the final grade submission deadline to submit projects. Students will be privately given project details via student email where they will have to acquire the data from specified sources. Course Assessment -- Assignments 20% 3 Exams for all modules 60% 3 Data Science Basics Quizzes 15% Will be precursors to exam(s) for the Data Science Basics module Make-up Student Project (being conditional) COURSE OUTLINE -- WEEK 1-3. Introduction to Generalised Linear Models (GLMs) with model estimation and summary statistics Multilinear Regression (fast fast review) Quantile Regression (fast review) Logistic Regression Poisson Regression WEEK 4-5. Hierarchical Modelling (HM) Introduction to HM Multilevel Modelling Random Effects and Mixed Models WEEK 6-7. Bayesian Statistics in Biostatics Note: I don’t introduce things to be a disgusting, miserable, viral bastard. Module will be extremely goal oriented, namely, problem, goal(s), methodology, logistics, implementation, evaluation. No social nor psychological probes/inquisitions upon students; there are certified licensed professionals elsewhere tied to meaningful or economic interests. Bayesian Inference (to the point, constructive and economical) Markov Chain Monte Carlo (MCMC) Methods – only constructive and economical methods Bayesian Regression Models WEEK 8-9. Advanced GLMs and Extensions Negative Binomial Regression Zero-Inflated Models Generalised Estimating Equations (GEE) WEEK 10-15. Data Science Basics Note: subjects of overfitting or underfitting arise in model validation statistics. Data Acquisition. Data Probing: glimpse(), str() Data Wrangling (functions from dplyr R package with piping) Summary Statistics, Skew, Kurtosis, Correlation Analysis and Heatmaps Machine Learning Overview Feature Selection with R (underlying methods may not be fully comprehended, but that’s generally the world): Principal Component Analysis (PCA), Kernel PCA, Boruta, FSelectorRccp. Comparative observation among such prior three also expected. Multiple Regression (very rapid review) OLS and Quantile Classification Logistic Regression Support Vector Machines Decision Trees Random Forests Clustering (K-Means and DBSCAN advanced repetition) Includes the Elboew Method, Silhouette Score and Davies-Bouldin Index Prerequisites: General Biology II, General Chemistry II, Biostatistics II Cell Biology: The standard definition of a cell in most introductory biology texts includes the line that cells are “the fundamental building blocks of all organisms.” Because of this fact, trite though it may be, a detailed understanding of the fundamental processes of cellular function is critical to all specialties within biology, clinical or academic. Some of these processes, including for example the biochemical mechanisms underlying cellular energetics, are remarkably consistent from bacteria to human. Other cellular processes and structures vary from cell type to cell type or organism to organism, allowing for unique adaptations of cells and organisms to particular functions. For example, nerve cells have various properties allowing them to conduct electrical signals and therefore process information, while kidney cells are specialized for the secretion of waste, and red blood cells for the transport of oxygen and carbon dioxide. What are the differences in physiology from cell type to cell type determining these specific functions? During the first half of the semester we will focus primarily on the biochemical processes that underlie cellular function, with an emphasis on protein structure and function, ion transport mechanisms and energy metabolism. The second half of the semester will emphasize more the function of particular organelles, including cell membranes, intracellular compartments and the cytoskeleton, and the relevance of these structures on processes like cell signaling and mitosis. Throughout the course, we will emphasize how variability in these processes imbues different cell types with their unique functional abilities. We will also seek to understand the experimental evidence for the different facts and concepts we study: How do we KNOW that nerve cell signaling, for example, involves the release of neurotransmitters? Some of this experimental evidence will be explored in a hands-on way in the lab sections, some will be discussed during lecture, and some will be the subject of analysis in the reading of original scientific manuscripts. Finally, we will examine how malfunctions in the cellular processes we are studying underlie certain diseases. In particular, the final few lectures of the course will focus on the biology of cancer cells: how do changes in cellular processes allow cancer cells to proliferate and metastasize? What are some of the current clinical approaches to curing cancer by blocking or reversing these processes? Aspirations --> To understand fundamental concepts of cellular function. To understand, and be able to critically analyze, the scientific evidence underlying our current understanding of cellular processes. To develop skills, through lab experiments, in some of the specific methodologies used in the study of modern cell biology. To become skilled at formulating and testing hypotheses using these methods. To develop a preliminary ability to read and analyze the primary scientific literature: What are the major findings of a science paper? What evidence is presented to support these findings? Are there shortcomings, either in the methods used or the logic of the experiments, which might lead one to question the conclusions reached by the authors? To be able to put this knowledge into larger contexts of how disease states occur or how organisms function adaptively within their environments. Typical text: The World of the Cell, by Becker, Kleinsmith, and Hardin, 6th edition (2006), Pearson/Benjamin Cummings Class Requirements and Grading --> 1. Class Participation (10%) To include attendance, responses to questions I pose in class, participation in discussions, and simply raising your hand from time to time to ask questions or make a comment (something I DO expect you to do). 2. Quizzes (10%) Two short in-class quizzes during the first half of the course. 3. Homework/problem sets (5%) There won’t be many of these; I’ll assign them when we hit subjects that are especially involved to help you learn the material and to make sure everyone is on track. 4. Primary literature readings (10%) We will read two papers from the primary scientific literature during the second half of the course. In both cases there will be an in-class discussion of the paper and a “reading guide” set of short essay questions which will be graded. The first reading guide assignment will be due AFTER the in-class discussion; the second assignment will be due BEFORE the in-class discussion. 5. Laboratory reports (25%) The specifics of each week’s lab report will be discussed during lab section. Typically, each week’s lab report will be due the following Monday in lecture. 6. Mid-term exam (20%) TENTATIVE format to include an in-class component, a short oral component, and a take-home component. 7. Final exam (20%) LABS --> Lab instructions for each week will be handed out ahead of time, either distributed as hard copies in lecture or posted on the course Angel site (or both). You are responsible for reading the instructions before lab. Otherwise, labs tend to run late, you will have difficulty obtaining the necessary data and knowing what to do with it. Do not expect the instructor to go over every step of the lab procedure before you start. Labs will make great emphasis on strong, practical and constructive immersion into the following software to accompany hands-on activity: << VCell, TiQuant + TiConstruct + TISIM >> Such software provides strong quantitative/computational microscopic assessment of specimens (or whatever) at professional standards. Such provides better means of objectives and expectations towards hands-on labs. Each lab will be associated with an explicit lab report assignment (contained in the lab instructions), usually due in lecture the Monday following lab. Usually, you may either submit the report with your lab partner or independently. If a report is submitted jointly, both partners must have contributed equally, as per Honor Code responsibilities. Do NOT make the mistake of dashing off reports the night before in a single draft. These reports will collectively account for 25% of your course grade, so take them seriously. The lab is a potentially dangerous place and you are required to follow all instructions given by your lab instructor and presented in the lab instructions. Disregarding instructions, or coming to class late or unprepared, may result in grade penalties, in addition to being just plain dangerous for yourself and those around you. Note: students can apply stationary video recording of labs with assigned regulations (to be given). Course Topics: Chapter 1 -19, 24. Some topics will require at least one week of instruction. Labs --> Cell Culturing, Aseptic Technique Cell Culture: basic techniques, population curve Cell Counting and splitting plates Cell Staining Histology Electron microscope Cell Harvesting & Cell Lysis Fractionalization of cells Common method(s) will be implemented Discussion and logistics for immunomagnetc separation & magnetic beads Isolation of erythrocyte membrane proteins Analysis of erythrocyte membrane proteins Bradford Assay (also identifying advantages and disadvantages) SDS-PAGE Chloroplasts and the Hill reaction Prerequisites: General Biology I & II Organic Chemistry I In-depth study of: (i) the structure of organic compounds and the functional groups (bonding, acid-base properties, nomenclature, conformations, stereochemistry), and (ii) the synthesis and reactivity (including detailed mechanisms) of alkanes, alkenes, alkynes, halides, alcohols, ethers, epoxides, sulfides and organometallic reagents. Laboratory experiments are related to topics covered in lecture and emphasize organic laboratory techniques, synthesis and spectroscopic characterization of organic molecules. Typical Texts: McMurry, John E. Organic Chemistry. 8th Edition. Brooks/Cole, 2012. McMurry, Susan. Study Guide with Student Solutions Manual. 8th Edition. Brooks/Cole. Typical Lab Manual: Barbaro, John and Richard K. Hill. Experiments in Organic Chemistry. 3rd Edition, Contemporary Publishing Company of Raleigh, Inc., 2006 Grading: 3 Exams (50% combined) Cumulative Final Exam (25%) Labs (25%) (On the occasion of significant improvement on the final exam, more weight will be placed on the final exam) INSTRUCTIONAL METHODS: List the different instructional methods you might use, in the course of the semester. List supplementary learning options, if any: Traditional lecture with use of chalkboard Computer assisted diagrams and graphics Molecular Models Team work in the laboratory Homework assignments Solving specific questions related to content studied Written exams and distribution of study questions/previous exams Use of the Internet UNIQUE ASPECTS OF COURSE (such as equipment, specified software, space requirements, etc.): Organic chemistry laboratories and their associated equipment, instruments and chemicals. Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Lecture Outline --> Ch. 1 Structure and Bonding Bonding; Hybridization; Drawing Chemical Structures; Functional Groups; Intro to IR Spectroscopy Ch. 2 Polar Covalent Bonds; Acids and Bases Chemical Bonding (Ionic and Covalent); Electronegativity and Dipole Moments; Formal Charges; Resonance Structures; Acid Base Theory (Bronsted-Lowry, Lewis); Acid and Base Strength (pKa); Acid-Base Reactions; Organic Acids and Organic Bases Ch. 3 Organic Compounds: Alkanes and their Stereochemistry Alkanes, Alkane Isomers, and Alkyl Groups; Properties of Alkanes; Conformations Ch. 4 Organic Compounds: Cycloalkanes and their Stereochemistry Cis-Trans Isomerism in Cycloalkanes; Stability and Conformations of Cycloalkanes; Chairs Ch. 5 Stereochemistry at Tetrahedral Centres Enantiomers, the Tetrahedral Carbon and Chirality; Optical Activity; R/S Sequence Rules; Diastereomers and Meso Compounds; Racemic Mixtures, Resolution of Enantiomers; Prochirality; Chirality in Nature Ch. 6 An Overview of Organic Reactions Kinds of Organic Reactions (Radical and Polar); Mechanisms; Describing a Reaction (Equilibria, Rates, Energy Changes, Bond Energy; Transition States, and Intermediates) Ch. 7 Alkenes: Structure and Reactivity Preparation and use of Alkenes; Cis-Trans Isomerism; Alkene Stereochemistry and E/Z Designation; Stability of Alkenes; Electrophilic Addition Reactions; Markovnikov’s Rule: Carbocation Structure and Stability; Carbocation Rearrangements Ch. 8 Alkenes: Reactions and Synthesis Preparation of Alkenes via Elimination Reactions; Addition Reactions of Alkenes (Halogenation, Hydration, Halohydrins, and Hydrogenation); Oxidation of Alkenes (Epoxidation and Hydroxylation); Addition of Carbenes; Radical Additions to Alkenes (Polymer Formation); Reaction Stereochemistry Ch. 9 Alkynes: An Introduction to Organic Synthesis Preparation of Alkynes; Addition Reactions of Alkynes (X2, HX, H2O, H2); Oxidative Cleavage; Alkyne Acidity and Alkylation; Introduction to Organic Synthesis Ch. 11 Reactions of Alkyl Halides: Nucleophilic Substitutions and Eliminations SN2, SN1, E2, E1, E1cB Reactions; Zaitsev’s Rule; Deuterium Isotope Effect Ch. 10 Organohalides Preparation of Alkyl Halides and Grignards; Radical and Allylic Halogenation; Organic Coupling Reactions, Redox in Organic Chemistry Ch. 17 Alcohols and Phenols Properties of Alcohols and Phenols; Preparation and Reactions of Alcohols; Reactions of Phenols Ch. 18 Ethers and Epoxides; Thiols and Sulfides Synthesis and Reactions of Ethers; Cyclic Ethers (Epoxides); Reactions of Epoxides: Crown Ethers; Thiols and Sulfides LABS --> Some experiments require more than one lab period to complete. Based on an instructor’s preference, availability of equipment/supplies or constraints within a given semester, this laboratory schedule is subject to change, including but not limited to, the addition or replacement of one or more of the above experiments with the following experiments: Addition of Bromine to E-Cinnamic Acid in Methylene Chloride Substitution Reactions of Alkyl Halides: Relative Rates Triphenylmethanol with Hydroiodic Acid 1. Check-in, Laboratory Safety, Practices and Waste Disposal. Simple Distillation. 2. Spectroscopy: Introduction to Infrared Spectroscopy. 3. Recrystallization, IR and Melting Point of benzoic acid. 4. Extraction of Organic Compounds from Natural Sources: Trimyristin from Nutmeg. 5. Paper Chromatography 6. Dehydration of Cyclohexanol. 7. Dimerization of 2-Methylpropene 8. Preparation of Diphenylacetylene Starting from Trans-Stilbene. 9. Preparation of Butyl Bromide/Preparation of t-Butyl Chloride (SN2/SN1). 10. Oxidation of Isoborneol to Camphor. 11. The Williamson Ether Synthesis: Preparation of Aryloxyacetic Acid from Cresol. Prerequisites: General Chemistry II Organic Chemistry II Evaluation of Performance --> There will be 1000 points possible in the course. Five exams will be given throughout the semester, consisting of four regular hour exams and a comprehensive final. Each of these exams will be worth 200 points, and your lowest score will be dropped. The four remaining highest scores will total 800 possible points. There will be 120 points possible from laboratory reports, 60 points possible from online D2L quizzes, and 20 points assigned to your laboratory notebook. Students will be kept updated on their performance throughout the semester. Laboratory --> Organic laboratory and lecture complement each other. The lecture supplies fundamental theory about molecular and electronic structure, chemical reactions, and their mechanisms. In the laboratory you will put this knowledge into practice to help you more fully understand the chemical process in progress. Typical text: Janice Gorzynski Smith "Organic Chemistry" with Solutions Manual, 3rd Ed. McGraw Hill Typical Lab Text: Pavia, Lampman, Kriz, and Engel "Techniques in the Organic Laboratory, Microscale and Macroscale", Harcourt College Publishing. There will be,specified software for use throughout course and labs. Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Course Outline --> Week 1 Mass spectroscopy. Infrared Spectroscopy Week 2 NMR: position of signals and strength of signals Spin-spin splitting, other 1 H NMR facts NMR: solving unknowns, 13C NMR Week 3 Reduction of alkenes, alkynes, R-X, and epoxides Epoxidation, dihydroxylation, oxidative alkene cleavage Oxidation of alcohols EXAM 1 Week 4 Radical reactions, alkane halogenation, chlorination of ethane Chlorination of other alkanes, bromination, allylic bromination Lipid oxidation, antioxidants, polymers Week 5 Resonance, conjugation, dienes Diene stability and the Diels-Alder reaction Week 6 Benzene nomenclature and structure Benzene’s unusual stability, criteria for aromaticity Aromatic rings of other types EXAM 2 Week 7 Halogenation, nitration and sulfonation Friedel-Crafts reactions, effects of ring substitution, limitations Disubstituted benzenes, side chain reactions, synthesis Week 8 Carboxylic acids: naming, properties, and preparation Carboxylic acid, reactions, and acidity Carbonyl chemistry: reductions of aldehydes & ketones Week 9 Reduction of carb. acid derivatives, organometallic reagents Synthesis, organometallic reactions EXAM 3 Week 10 Aldehydes and ketones: naming, properties, preparation Aldehyde/ketone reactions, nucleophilic addition, Wittig rxn Wittig rxn continued, imines Week 11 Acetal formation/hydrolysis/use in protecting C=O Carboxylic acids and their derivatives Reactions of acid chlorides, anhydrides Week 12 Reactions of carboxylic acids, esters, and amides Summary of acyl substitutions, applications Keto-enol tautomerism; the aldol reaction EXAM 4 Week 13 Review for final exam Week 14 comprehensive final exam LABS --> Spectroscopy, no lab Exp. 1 FTIR, MS: Check in. Isolation and characterization of Eugenol (essence of cloves) Exp. 2 MS, IR, NMR: Spectral Identification of Organic Compounds Exp. 3 Oxidation: Oxidation of Cyclohexanol to Cyclohexanone Exp. 4-1 Spectroscopy: Identification of a General Unknown, part 1 Exp. 4-2 Spectroscopy: Identification of a General Unknown, part 2 Exp. 5 Diels-Alder Reaction: Synthesis of 4-Cyclohexene-cis-1,2-dicarboxylic Anhydride Exp. 6 Aromatic Substitution: Electrophilic Aromatic Substitution: the Nitration of Toluene Exp. 7 Carbonyl chemistry: Reduction of Heptanal Using Sodium Borohydride Exp. 8 Organometallic Reagents: Preparation & Carbonation of a Grignard Reagent: Benzoic Acid Exp. 9 Carbonyl chemistry: The Synthesis of an Alkene Using a Wittig Reaction Exp. 10 Carboxylic Acid Derivatives: Flavours & Fragrances: Isopentyl Acetate synthesis (Banana Oil Exp. 11 Enolate Chemistry: Synthesis of 2 Methyl-2-pentenal --> An Aldol Condensation Check out Prerequisite: Organic Chemistry I Organic Synthesis Laboratory Practice of organic laboratory techniques. Three hours of laboratory per lab session, twice a week. Approved chemical safety goggles meeting whatever national standards. The purpose of this laboratory course is to introduce students to the techniques that organic chemists (as well as biochemists, physical chemists, etc.) use in their daily routines. After learning and understanding those techniques, students will apply their knowledge to new situations to understand synthesis reactions, molecular structure determination, and analysis of (un)known compounds. Organic chemistry laboratory is important for several reasons. It introduces students to many different laboratory practices and concepts that will be used in subsequent chemistry laboratory classes and in other laboratory situations in biology, pharmacy, and chemical engineering (just to name a few!). It is anticipated that by the completion of this course, students will be familiar with all of the following topics and techniques: Safety in the laboratory Interpreting and following scientific directions Keeping a proper lab notebook Names and proper usage of lab instruments Understanding of general properties of compounds (including solubility, miscibility, acid/base chemistry, etc.) Proper usage of glassware Isolation and purification techniques (including filtration, solvent removal, drying solutions, distillations, chromatography (thin-layer, column, and gas) and crystallization/recrystallization) Characterization techniques including spectroscopy and melting point determination Interpretation of scientific results including percent yield and recovery, melting point, boiling point, IR and NMR spectra, and Rf values Required Materials: A laboratory notebook with carbon(less) pages Approved safety goggles Lab coats Lab manual will be posted through Blackboard Typical text: C.F. Wilcox, M.F. Wilcox, "Experimental Organic Chemistry, A Small-Scale Approach", (3rd edition, 2010). Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Lectures --> Lecture sessions are designed to clarify the concepts covered in the lab, as well as give an overview of techniques that will be used in the lab. Attendance is expected: The labs are only 3 hours in duration, so these lectures will be where you learn everything that you’ll need. Lab exercises will be available on Blackboard for each week. Please be considerate of your fellow students during the lecture period. Disruptions of any kind will not be tolerated and may result in expulsion from the classroom. Laboratory --> You will be required to have appropriate clothing before being allowed to enter the lab. Pre-labs are due at the beginning of the lab, and results and postlabs are due at the beginning of the lab 1 week after completion of the experiment! You will be expected to adhere to all of the lab safety rules. You are all expected to do your part to maintain a clean lab environment as part of GLP (Good Lab Practices): All reagent and solvent bottles should be completely closed immediately after use; All spills and dribbles should be cleaned immediately; All glassware should be put away at the end of the lab, and walkways should be kept free of debris. The following is the distribution of possible points in the course: Library Searching Exercise Database Search Exercises (Spectroscopy and Chromatography) Lab Quizzes Reaction/Synthesis methods knowledge Appropriate choice of method Appropriate constituents and tools. Procedure/steps (summary and/or ordering) Stoichiometry problems Spectroscopy and/or Chromatography analysis/interpretation Applications and industries Multistep Reaction/Synthesis Labs Lab Cleanliness Pre-lab Submissions Lab Notebook and Reports Lab Final Day 1: Much resemblance to quizzes Day 2-3: Augmented with the following: Molecular modelling software exercises Two or Three Practicum Group Labs (open notes) Part A. Points deducted for incompetent questionnaire for safety procedures for respective lab Part B. 2-3 labs to be implemented with competent data recording and lab reports. YOUR LAB REPORT CONSISTS OF THREE (3) PARTS --> Part I - Prelab Report. A copy of your lab notebook pages containing the lab write-up and answers to any prelab questions. This is due at the start of each experiment. Part II - Results. A copy of your notebook pages containing observations noted during the lab experiment. Is due with Part III one week from the conclusion of the experiment. Part III - Postlab Report. A summary of results and answers to postlab questions. This can be written on separate loose-leaf paper. Is due with Part II one week from the conclusion of the experiment Course Outline: Week1 Check-in/Safety Video/ Safety Procedures and Regulations Fractional Distillation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation, results and analysis Week 2 Measuring the Melting Points of Compounds and Mixtures Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 3 Purification by Recrystallization and Melting Point Measurement Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 4 Nucleophilic Substitution: Synthesis (SN1 Mechanism and SN2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 5 Oxidation of Alcohols (Primary, Secondary and Tertiary). Infrared Spectroscopy. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Infrared Spectroscopy Results and analysis Week 6 Elimination Reaction (E1 Mechanism and E2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 7 Synthesis of Aspirin. Chromatography and/or Spectroscopy Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Chromatography and/or Spectroscopy Results and analysis Week 8 Solvent Extraction Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 9 Electrophilic Aromatic Substitution: Synthesis of o- and p-Nitrophenol. No distillation; extract product with ethyl acetate. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 10 Separation and purification of o- and p-Nitrophenol by Liquid Chromatography. Use 100 mg sample, check by chromatography. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Results and analysis Week 11 Aldol Condensation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 12 Grignard Reaction: Synthesis of Phenylmagnesium Bromide. Week 1: Part 1. Add methyl benzoate and sustain the desiccator for next week. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 13 HCl workup of previous week’s product. Synthesis of Triphenylmethanol and recrystallization of product. Purity check by melting point measurement. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 14 -15 Wrapping up/cleaning things up. Final Exam. Prerequisite: Organic Chemistry I Biochemistry: The study of biochemistry investigates the interplay between biological macromolecules such as proteins and nucleic acids, and low molecular weight metabolites (such as the products of glucose metabolism). In this course, you will apply your knowledge of intermolecular forces, thermodynamics (when a reaction occurs), chemical kinetics (how fast a reaction occurs), and chemical structure and functionality to understand how biological molecules (and life) work. COURSE GOALS AND OBJECTIVES (Our Roadmap!) -Be able to describe/identify the forces that direct/stabilize different levels of protein structure -Be able to predict how changes in amino acid (or nucleotide) sequence can affect macromolecular structure and function -Be able to explain how enzymes are able to affect reaction rate enhancement -Be able to articulate and apply what the enzyme parameters of KM, Vmax, kcat and kcat/KM tell us about an enzyme -Be able to describe the interactions of biomolecules both quantitatively and qualitatively (in many cases, including mechanistic details) -Be able to understand the flow of metabolic intermediates through a pathway and communicate information about metabolic pathways using diagrams -Be able to describe multiple experimental methods used in biochemistry, interpret data from these methods to form conclusions, and develop a testable hypothesis to answer a question -Be able to summarize and analyse primary literature and data, and apply gathered information to new situations -Increase problem solving skills such as: critical thinking, data analysis, graphical analysis -Increase process skills such as: communication of scientific concepts and experimental results, group dynamics and teamwork, management and self-assessment -Develop a community of active learners who are intentional about their educational choices Course Materials: Calculator Emphasis on reinforcing skills with software --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Typical Texts --> Nelson DL and MM Cox. Lehninger Principles of Biochemistry (5th edition). (“Lehninger”) Loertscher J and V Minderhout. Foundations of Biochemistry (3rd edition). (“FOBC”) Additional --> Blast protein databases, align protein sequences, build protein homology models, and evaluate the quality of these models Lab Manual examples --> Lasseter, B. F. (2020). Biochemistry in the Lab: A Manual for Undergraduates. CRC Press Related to week 6: Edwards, P., Zhang, C., Zhang, B. et al. Smartphone based optical spectrometer for diffusive reflectance spectroscopic measurement of hemoglobin. Sci Rep 7, 12224 (2017). Course Overview --> You will frequently be given initial assignments to work on as an individual before class. These assignments must be ready at the start of class – your preparation will form part of your weekly participation grade. During our class meeting time, you will frequently function as a member of a Learning Team, developing and examining chemistry concepts as a unit. Your team effort and participation is part of your weekly participation grade. The team responses to a few Key Questions on each in-class activity will be evaluated for strength of concept and effective communication of the concept. The team will also strategize on ways to improve teamwork and team products. These responses will also form part of your weekly participation grade. Application exercises will be assigned for each activity. Together with problems from the text, they will form your weekly problem set that will be collected and graded for each individual. These homework problems and exercises are important to your success in the course. Actively working these homework problems is essential for your understanding of the material, as they bring your concept development full circle. The questions will be drawn from lectures, in-class activities, problem sets and discussions, as well as relevant primary literature that you may not have been previously assigned. The purpose of doing biochemistry is to gain experience in experimental methods that you’ll be reading about throughout the semester. Attendance on your scheduled lab day is expected. Software activities concerning biochemistry will accompany labs. Software activities concerning biochemistry will accompany labs as pre-lab development or post simulations. Grading --> Team Participation Problem Sets/Other Laboratory 2 Midterm Exams Final Exam Lecture Outline --> Week 1 Introduction to Biochemistry Week 2 Intermolecular forces and water. Amino acids and peptide bonds Week 3 Protein Folding Week 4 Working with proteins Week 5 Enzyme catalysis. Enzyme Kinetics Week 6 Enzyme inhibition. Hemoglobin Week 7 Exam 1; Carbohydrates Week 8 Glycobiology Week 9 Lipids and membranes. Transport across membranes Week 10 Signal transduction. Metabolism overview Week 11 Glycolysis. Glycolysis regulation and related pathways Week 12 Glycogen metabolism and gluconeogenesis. Citric Acid Cycle Week 13 Electron Transport Chain / Oxidative Phosphorylation; Exam 2 Week 14 Lipid metabolism. Nucleotides and nucleic acids Week 15 Nucleic acids structure and function Week 16 Final Exam Prerequisite: General Biology I. General Chemistry I & II. Co-requisite or Prereq: Organic Chemistry I Food Chemistry Water, carbohydrates, lipids, proteins, vitamins, and minerals in foods; biochemical and functional properties, enzymes, food additives (emulsifiers, pigments, colours, flavours, preservatives, and sweeteners) and texture as related to properties in food systems and during processing. Students will be able to identify the structure of food constituents, relate this structure to the constituents’ function and describe the constituents’ roll in respect to food quality, nutrition, safety, processing, etc. Students will also be able to differentiate chemical interactions, reactions of food components, their effect on sensory, nutritional and functional properties of foods, and how processing influences these reactions. The student will be able to explain how environmental factors such as temperature, pH, ionic characteristics and strength, bonding, light, etc. effect chemical changes in food systems and be able to adjust these conditions to improve or minimize chemical and biochemical deterioration of food systems. Finally, the student will be able to integrate chemistry and biochemistry principles into real-world food science and nutritional problems. Typical text: Food Chemistry, 4th Edition. 2008. S. Damodaran, K.L. Parkin, O.R. Fennema Eds. CRC Press Supplemental Reading: Introductory Food Chemistry, 2013. John Brady, Cornell University Press, New York. Typical Texts for Labs: The Food Chemistry Laboratory: A Manual for Experimental Foods, Dietetics, and food Scientists, Second Edition (Contemporary Food Science), CRC Press Food Chemistry: A Laboratory Manual. John Wiley & Sons, by Dennis, D. Miller. A total of three (3) exams will be administered over the course of the semester during the lecture period. There is no comprehensive final for this class. Each exam will be worth 13% of the final grade. The exams will be taken in “Blackboard” and you must have the Lockdown Browser downloaded onto your laptop prior to the first exam. NOTE: lab activities will run for 15 weeks; particulary for experiment 9 to acquire good data. Else, such additional time can serve as makeup for cancelled days or emergencies. Evaluation Method --> All assignments and answers on exams will be expected to be of professional quality. Laboratory reports are due generally two weeks after the lab exercise or as noted on the lab syllabus. No late assignments will be accepted without prior approval from the instructor. 3 Exams 39% Attendance and participation (lecture) 11% Laboratory 50% Course Outline: WEEK 1 (Chapter 2) Introduction. Review. Water. Water activity WEEK 2 (chapter 13, Chapter 3) Gels, emulsions, foams. Carbs intro. WEEK 3 (Chapter 3) Carbs: Starch. Other Carbs. WEEK 4 (Chapter 14, Chapter 4) Browning Reactions. Lipid components EXAM 1 WEEK 5 (Chapter 4) Review of lipid components. Lipid properties. Lipid process, functionality WEEK 6 (Chapter 4) Lipid deterioration WEEK 7 (Chapter 5) Amino acids, protein structure. Protein denaturation & functionality WEEK 8 (Chapters 5, 15, 16) Protein processing & modification. Milk & meat proteins EXAM 2 WEEK 9 (Chapter 6) Enzymes WEEK 10 (Chapter 7, Chapter 8) Vitamins. Minerals WEEK 11 (Chapters 9, 10, 11) Pigments and colorants. Flavors. Additives WEEK 12 (Chapters 11, 12, 17) Additives-sweeteners & fat replacers. Bioactive compounds. Plants EXAM 3 LABS --> First of all, food chemistry experiments very often do not work out as we planned. Chemicals are not always pure, the conditions are not extraordinarily controlled, and you could not reasonably be expected to get a “right” or “perfect” answer. Else, something is wrong with the way whatever works. In food chemistry we often have poorly characterized starting materials, food ingredients, and many reactions occurring in parallel under non-ideal conditions. Unsurprisingly the data we get is often noisy and hard to interpret. Wherever you go in life you will be trying to make difficult business decisions based on poor data. Developing these analytical skills in this this class will certainly benefit you as a manager in the food industry. Students should be able to: 1. Recognize the important reactions in food chemistry and their consequences. 2. Be familiar with methods to measure these reactions. 3. Be capable of reporting their results in an appropriate format. 4. Be capable of designing and conducting an experiment to understand a simple food chemistry problem. Lab Scoring --> Pre lab questions (12 labs x 20 points) Laboratory Participation (12 labs x 10 points) Laboratory Reports (9-10 reports x 25 points) Laboratory Notebooks (9-10 labs x 5 points) Note: each lab’s duration will be 1 week (but special case is experiment 9). Experiment 1 Physical Properties of Foods: water activity, specific gravity, viscosity and refractive index Experiment 2 Dispersions of matter: solutions, emulsions, and foams Experiment 3 Ice crystal formation Experiment 4 Carbohydrates: reducing sugars, starch morphology, and gelatinization Experiment 5 Lipid characteristics Experiment 6 Proteins: qualitative and quantitative analysis Experiment 7 Proteins: Maillard browning, effect of heat Experiment 8 Enzymatic browning Experiment 9 Fermentation: concern will be using different extracted fruit juices towards the most important phase in wine making (being fermentation). A clearly designed fermentation system will be developed and constructed. Fermentation system will have the following elements w.r.t. time: Means of determining carbon dioxide content in atmosphere Means of determining oxygen Pressure sensing Means to extract samples at different designated periods (hopefully not compromising process) Temperature sensing w.r.t. time Verification of “sum of all parts” at final stage and explanation for any possible discrepancy Will have 2-3 different types of fruit extractions Comprehensive knowledge of composition of fruit extract Average Sugar content level (accounting for different types) For the fermentation agents there will be three circumstances Without any added, sugar, water and yeast Added sugar and water Adding yeast solely Adding yeast, sugar and water For such fermentation agents type and quantity must be known. Namely, type of sugar (or honey), scientific identification of yeast type. As well, amounts to be added. Note: different benign bacteria may also come into play with a type of fermentation that’s unique to fermentation from yeast. Based on all such prior data detailed description of of metabolic processes will be acquired, which will emanate detailed biochemical processes along with the stoichiometry. Determination of microbial ecology at different stages. What type of alcohol will be produced? Can chemical process be developed analytically to give an instantaneous mathematical model for alcohol production? Determination of alcohol content and other by-products (different acids and glycerol) at different stages. Determination of ecology at end date. Note: temperature studies, and studies among various added yeast and added sugar agents may or may not fit time schedule. Note: fermentation towards yogurt can also be done as alternative or compliment, but with appropriate agents, environment construction, analysis etc. Software of interest R + R Studio Excel COCO (+ ChemSep), DWSIM (+ ChemSep) << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> Note: data modelling and data analysis expected (both deterministic and statistical). Experiment 10 Food flavours and precursors Experiment 11 PART A (ripeness) Das, A. et al. (2016). Ultra-Portable, Wireless Smartphone Spectrometer for Rapid, Non-Destructive Testing of Fruit Ripeness. Sci Rep 6, 32504 Note: not interested in the additive manufacturing part, rather assembling primitive prototypes out of the components to implement. Note: generally use fruits attainable in your environment as substitutes. Side component 1: It’s important to characterise what type of molecules, polymers, etc. are distinctively prevalent for ripeness (for respective fruit). Considerable variation in freshness, say, groups consisting of consumable U were stored (G °C, I% RH) for 0, 2, 4, 6, 8, 10, etc., etc. days. Apply different types of consumables. Side component 2: Experiments to conduct alongside side component 1 involving the same fruit speciments with 0, 2, 4, 6, 8, 10, etc., etc. days. Use fruits attainable in your environment as substitutes:-> Montero, T. M. et al (1996). Quality attributes of Strawberry During Ripening. Scientia Horticulturae, volume 65 Issue 4, pages 239 – 250 Ninio R. et al. (2003). Changes in Sugars, Acids, and Volatiles During Ripening of Koubo [Cereus peruvianus (L.) Miller] Fruits. J Agric Food Chem. 29; 51(3): pages 797-801 Anthon GE, LeStrange M, Barrett DM. (2011). Changes in pH, Acids, Sugars and Other Quality Parameters During Extended Vine Holding of Ripe Processing Tomatoes. J Sci Food Agric. 91(7): 1175-81. PART B (staleness) It’s important to characterise what type of molecules, polymers, etc. are distinctively prevalent for staleness (for respective consumable). Considerable variation in “age”, say, groups consisting of consumable X were stored (A °C, B% RH) for 0, 2, 4, 6, 8, 10, etc. etc. days. Apply different types of consumables. Note: generally use fruits attainable in your environment as substitutes. Note: there can be side component 2 like what is done in part A. Experiment 11 Integrative activity: Nutrient food party (pyramid challenge) Experiment 12 Integrative activity: Nutrient food party (pyramid challenge) Prerequisites: Biochemistry, Organic Chemistry I Genetic Engineering & Technology Course textbook: Molecular Biotechnology: principles and applications of recombinant DNAB.R. Glick and J.J. Pasternack, 3rd edition 2003 Course Grade Constitution --> 3 - 6 Assignments 3 Exams Course Outline --> Week 1 Introduction-Genetic Engineering and a tour of Genome Space DNA/RNA Processing and Gene Expression (a review): Week 2 Basic Techniques of Molecular Biology Restriction Endonucleases, Vectors, Cloning Week 3 Library Screens PCR, Sequencing Week 4 Exam Review and Exam Week 5 Prokaryotic Gene Expression Eukaryotic Gene Expression Genetic Engineering in Plants I Week 6 Genetic Engineering in Plants II: Applications Genetic Engineering in Plants III: the Next Generation of Rice Week 7 Sequence Analysis, Genome Structure Comparative Genomics Week 8 Functional Genomics: Analysis of Gene Expression Modifying Gene Expression and Cellular Function Week 9 Exam review and Exam Week 10 OPV and the emergence of AIDS The Origin of AIDS, con’t; vaccine intro; Edible vaccines Vaccine targets: malaria and ebola Week 11 Human Gene therapy I Human Gene therapy II: examples Week 12 Genetic engineering in animals I: Cloning Genetic engineering in animals II: Knockouts and Knockins, Inducible Gene Targeting Week 13 Genetic engineering in animals III: examples Genetic engineering in animals IV: xeno transplantation part a Genetic engineering in animals IV: xeno transplantation part b Week 14 Ethics and Patent Law Week 15 Exam Review and Exam Comparative Genomics An introduction to the concepts and experimental approaches used in microbial genomics. The laboratory will allow students to familiarize themselves with software routinely used in genomics and proteomics. Although the focus of the material is mainly on microbial genomes many of the approaches covered in the class can be applied to any system. An introduction into eukaryotic genomics is also provided. Literature: A Primer of Genome Science. Second Edition. Gibson and Muse. Sinauer Associates. Software --> << COPASI, Pathvisio + Cytoscape + KEGG >> RStudio and R packages for genome data development (BiocManager, ChemmineR, dPCP, MAPITR) Thodberg, M., & Sandelin, A. (2019). A step-by-step guide to analysing CAGE data using R/Bioconductor. F1000Research, 8, 886 Cao, Y., Charisi, A., Cheng, L. C., Jiang, T., & Girke, T. (2008). ChemmineR: a compound mining framework for R. Bioinformatics (Oxford, England), 24(15), 1733–1734 < manuals.bioinformatics.ucr.edu/home/R_BioCondManual > < cran.r-project.org/web/packages/BiocManager/vignettes/BiocManager.html > NOTE: will like to incorporate the R environment into labs as compliment to analyses. Assessment --> 3 – 4 Quizzes - 120 points Laboratory – 75 points (problem sets and software) Two exams - 100 points each. Final lab project - 50 points Group Projects - 75 Labs --> Lab reports are due ONE WEEK after the scheduled lab time. Group Projects --> 1. Students will assemble, annotate and describe a mock genome project using public and/or simulated sequence data. 2. Genome confirmation of various samples (from both animalia and plantae) for DNA sequencing the lab. Student groups with collect samples of at least 3 elements of animalia, and at least 3 elements of plantae. Will have lab DNA sequencing activities towards identification of genomes; identify possible using genetic markers to compare with databases. For animalia samples preference to be less sophisticated organisms, say, small genomes due to time constraints. NOTE: for the microbiology students lab methods will come from the following for microbial samples L. Barth Reller, Melvin P. Weinstein, Cathy A. Petti (2007). Detection and Identification of Microorganisms by Gene Amplification and Sequencing, Clinical Infectious Diseases, Volume 44, Issue 8, Pages 1108–1114 Fraser, C., Eisen, J. & Salzberg, S. (2000). Microbial Genome Sequencing. Nature 406, 799–803 C. Bertelli, G. Greub. (2013). Rapid Bacterial Genome Sequencing: Methods and Applications in Clinical Microbiology. Clinical Microbiology and Infection, Volume 19, Issue 9, Pages 803-813 Lasken, R. S., & McLean, J. S. (2014). Recent Advances in Genomic DNA Sequencing of Microbial Species from Single Cells. Nature Reviews. Genetics, 15(9), 577–584 Course Outline --> Week (1) Introduction to Genomics Microbial Genomes/Microbial databases Week 1 Lab – Introduction to lab/NCBI Week (2) Genomic webtools Sequencing and Assembly Week 2 Lab – Exploring web based tools for bioinformatics Week (3) Searching databases for homologs Tools for monitoring genome-wide gene expression Week 3 Lab – Sequence assembly Week (4) DNA microarrays DNA microarrays Week 4 Lab – BLAST & PSI-BLAST DNA microarraysProteomics Week (5) Structural Genomics Genome scale protein/protein interactions Week 5 Lab – DNA Microarrays/ Image analysis/data Week (6) High throughput genetics Comparative Genomic Week 6 Lab – Proteomics Week 7 Lab – ORF finder/genome annotation Week (9) Annotation Project/Exam Review of past exam Phylogeny of life Week 9 Lab – Lateral Gene Transfer Week (10) Molecular evolution and gene transfer Sequence alignment and phylogenetic analysis Week 10 Lab – Phylogeny Week (11) Systems Biology Genome based diagnostics Week 11 Lab – Begin Annotation project Week (12) Phylogenetic analysis (continued) April 7 – Metagenomics Week 12 Lab – Codon Bias Week (13) Eukaryotic Genomes Human Genome Project Week 13 Lab – Human Genome Project Week (14)SNPs Week 14 Lab – Annotation Project Week (15) Organellar Genomes Review Week 15 Lab – Annotation Project Prerequisites: Biochemistry, Organic Chemistry I, Genetic Engineering & Technology Molecular Biology I This Molecular Biology course is designed to give a good background in current Molecular Biology, which should allow for easy continuation to graduate or professional school courses. The major themes are Eukaryotic and Prokaryotic DNA replication, Chromosomal structure and function and Gene structure and function. Students will learn from current papers in the scientific literature, and will be expected to use concepts developed in the course in class, in the laboratory and in exams. Molecular Biology is fundamental to the study of all living things. It describes, in its most basic form, the mechanisms of how organisms live, reproduce and evolve. It is basic to much of modern Biology, no matter what the field of study. The purpose of this semester in Molecular Biology is to familiarize the student with those concepts that are basic to the functioning of prokaryotic and eukaryotic cells. Lecture is the foundation of the course. Laboratories will not always coincide with the lecture topics. The student is responsible for assignments that add to the lecture and lab material. The student is encouraged to seek out related materials that are available, such as scientific journals (e.g. Cell, Nature, Scientific American), newspapers, magazines and television programs (e.g. channels 12 and 52) that relate to course topics. Lecture notes will be published on the internet at my home page before the given lecture. There are three basic concepts in this course - the replication of DNA; the structure and function of chromosomes; and the structure and functioning of genes. They will be organized as indicated below. Students will reinforced with Molecular Biology laboratory techniques (DNA isolation and purification, recombinant DNA synthesis and cloning, gene detection, PCR and Southern and Western Blotting), which will be used to expand the student's appreciation and knowledge of the lecture material. The lectures cover Molecular Biology as a whole - the "central dogma" of Biology: DNA makes RNA which then makes protein. Out of this there arise three concepts - Eukaryotic and Prokaryotic DNA biosynthesis; Chromosomal structure and function (with associated proteins and functions); and Eukaryotic and Prokaryotic gene structure and function (mRNA, tRNA synthesis and function, including protein synthesis), and how they relate to basic biological and chemical concepts (such as the action of evolutionary processes on living things) learned in previous courses. In general, they should understand how our genomes function, including gene activation and deactivation, RNA synthesis and protein biosynthesis and be able to use this knowledge in their work and in the laboratory. Overall, emphasized and reemphasized in the course, and illustrated by specific examples and laboratory experiments, are the ways in which the above topics are interconnected, and factors used in one way are recycled to be used in another. This leads to interconnectiveness amongst the various cellular functions, and allows for signaling and controls between them. These principles should allow them to establish a firm connection between this course and other aspects of biology and give a foundation for future Molecular Biology courses and/or a good appreciation of concepts needed to make reasoned choices in their everyday lives. Typical Text: Watson et. al. Molecular Biology of the Gene ed. 5 Typical Laboratory Manual Text: Human Molecular Biology Laboratory Manual - S. Surzycki The professor's evaluation of student participation in lecture and laboratory can be used to benefit hard working students and possibly enhance their grade if they are in a borderline position. The laboratory grade is based mainly on the laboratory paper (normal scientific format, aprox. 8 - 10 pages, with a bibliography and internal referencing), as well as the instructor's assessment of the student's activity for the entire laboratory. Some laboratory based questions will appear on exams, especially including the final exam. Grading: Exams -15% of final grade (x 3 exams): 45% Laboratory -25% of final grade Final Exam -30% of final grade Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well to this course that further encourages a modern and profession environment, extending beyond memor based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities on e is interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Lab Outline --> Chapter 7 - Determination of Human Telomere Length (pages 164 - 195) All activities in chapter 7 must be done Chapter 8 - RT-PCR of Human Genes (pages 196 - 214) All activities in chapter 8 must be done Course Outline --> SECTION ONE: DNA replication, repair and recombination Chapters 1 – 11 SECTION TWO: Chromosome structure and function, chromatin, prokaryotic operon structure and function Chapters 7 & 16 SECTION THREE: The eukaryotic operon structure and function, gene clusters, genes in organelle Chapters 2, 3, 17 & 18 SECTION FOUR: Ribosomes, protein biosynthesis and transportation, eukaryotic and prokaryotic viruses, genetic engineering Chapters 14 & 16, 17, 13 LAB REPORT DUE & FINAL EXAM Prerequisites: Cell Biology, Organic Chemistry I Molecular Biology II Molecular Biology is a wide term encompassing two often complementary fields of study: a) laboratory and computer-based tools that can be used to study gene and genome identity and function (“molecular tools”), and b) the underlying fundamental structure of DNA and RNA. In this class there will be about a 50:50 split between focus on molecular tools (techniques) on the one hand, and the structure and function of DNA and RNA on the other hand. Proteins are often the focus of Biochemistry classes. Molecular Biology stresses the application of advanced molecular tools in the lab, and during analysis of scientific data presented in primary literature, in addition to covering genetic topics in more detail than was done in previous classes. The major expectations of students are : • To be become familiar with molecular techniques. • Understand the use of these techniques in the discovery of DNA and RNA metabolism and function. • Become more proficient at reading and critiquing primary literature. • Become familiar with commonly used laboratory techniques in the lab during an allsemester long research project. • Emphasis is not on memorization of the details of the molecular machinery of the cell. Instead, it is on developing skills to apply the learned techniques to the understanding of scientific discovery (data interpretation), as well as to suggest ways to study the function of molecules (experimental design). Molecular research is impossible to conduct in set 4-hour increments once a week. Typical Textbook/Readings: • Burton Tropp: Molecular Biology, 4th edition, 2012 Parts of some chapters will be used in this book in lecture, and book will be a good resource for looking up details, reading ahead or after class. Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well to this course that further encourages a modern and profession environment, extending beyond memor based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities on e is interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Grading --> 2 Midterm exams (each 70 points) 140 points 5 Quizzes (4x10 points each, quiz 5 =20 points) 60 points 1 Research paper (lab write-up) 40 points Lecture participation 20 points Carefulness in lab and preparation for labs 20 points Lab prelabs/postlabs ~30 points 3 Paper discussion prep and participation (4x20 points + 2x10 points) 100 points LABS --> It’s important to keep a neat and bound notebook for the lab, but you do not need to buy an expensive lab notebook. At the end of the semester you will be required to turn in a research paper describing the work you did on the lab project and put it in context of a greater scientific question. Only one lab report/paper will be handed in for the semester. This report reflects multiple weeks of work. Therefore make sure to collect gel photos, instrument readings, microarray and RNA-seq data analyses, so that you have all the files you need for your final paper. While all class and lab assignments have to be written individually unless specified differently, the final lab report can be written collaboratively with your lab partner, or individually if you prefer. Preparation for labs: Labs in this class are very expensive and it is of great importance that you come prepared to lab. In the past I have simply relied on suggesting that everyone read the manual before lab. However, it only takes one un- or underprepared person to make an experiment fail – often for more students than just that one person. So I have decided that some form of enforcement of the preparation requirement is needed and have reverted to prelab assignments, which test your level of preparedness. In addition there is now a grade for “carefeulness in lab”. This grade is based on whether you are prepared for lab or excessively ask unnecessary questions that you could have answered yourself by reading the manual. I do encourage asking questions, but I also encourage self reliance and careful attention to detail. Not every experimental failure is due to operator error. Such failed experiments are common and will not influence your lab grade negatively. Again, software mentioned earlier will accompany labs. Outline Week 1: Nucleic Acid Structure, Genome Organisation Week 2: RNA techniques (hybridization, reporters) RNA techniques (qPCR) DNA sequencing methods (Sanger) DNA sequencing methods (whole genome approach) Lab1: Microarray analysis - RNA Extraction Week 3: QUIZ 1: RNA techniques DNA sequencing methods (whole genome approach) Paper Discussion 1 Lab1: Microarray analysis c - DNA production Week 4: Gene mapping, map based cloning Human genome variation, the concept of “race” Lab1: Microarray analysis - Slide hybridization Week 5 Paper Discussion 2 - Human genome variation DNA Damage Quiz 3 (mapping/cloning) Lab1: Microarray analysis Statistics of microarray analysis Slide analysis: first part in lab. Finish slide analysis/statistics on your own time after all slides are pre-analyzed after [whenever] Week 6: DNA repair, Technique: EMSA (for paper 1) Paper Discussion 3 (DNA repair) Lab 2: RNA-seq analysis - Computer workshop uisng iPlant tools (or by software provided) Week 7: Recombination Lab 2: RNA-seq analysis - Analysis of RNA-seq data from week 1, and comparison with array data Week 8: Transposons Paper Discussion 4 (Recombination) Lab 3: Genetic marker analysis Sequence analysis using Genome Browser Primer design CAPS search Week 9: Eukaryotic transcriptional regulation Lab 3: Genetic marker analysis - DNA extraction, PCR set up Week 10: Epigenetics (technique: Immunoprecipitation) Paper Discussion 5 (Epigenetics ) Lab 3: Genetic marker analysis CAPS digest gel analysis (on your own) Lab 4: RNAi cloning - PCR of insert Week 11: QUIZ 4: The transcription unit RNAi Lab 4: RNAi cloning PCR clean-up and cloning reaction Transformation (on your own) Week 12: Paper Discussion 6 (RNAi) Lab 4: RNAi cloning - Finish up from week 11 Week 13: Splicing Agrobacterium and gene transformation Lab 4: RNAi cloning Colony PCR of cloned insert Glycerol stocks and sequencing of positive clones Week 14: Agrobacterium and gene transformation QUIZ 5: (molecular techniques) Lab 4: RNAi cloning Finish up from week 13 Final Exam Lab report due Prerequisites: Genetic Engineering & Technology, Molecular Biology I
Metabolism: Selected topics in metabolic pathways associated with carbohydrates, proteins, nucleic acids and lipid metabolism. This will include biosynthesis and degradation, cellular function, and their regulation and their physiological relevance. Additional topics will include hormones signalling pathways; cell signalling pathways and their metabolic roles and regulation. Emphasis will be given on the physiological relevance on different metabolic pathways including various clinical correlations. Direct examples from current literature will be given on a regular basis. Students who have successfully completed this course will be having thorough understanding on various metabolic pathways and their regulation and impact on physiological function and human health. Student should have fundamental knowledge on cell signalling and their regulatory mechanism. Emphasis will be given towards human health and disease correlation. Typical Text: Biochemistry with Clinical Correlations by Thomas M Devlin, 7 th Edition. Please note that additional study materials will be taken directly from current literatures and other resources. BIOCHEMICAL SOFTWARE --> Concerning particular topics and biochemical processes students will investigate with choice(s) out of the mentioned software below. Will have extensive observation and description of properties, characteristics of the processes, bonds, etc. in terms of organic chemistry and biochemistry. Each software activity goes along with a report. Multiple tasks given to each student (group). MUST connect with predominant or modern research, hence references from journal articles, etc. Each report to be 5-12 pages --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> ESSENTIALS --> << COPASI, Pathvisio + Cytoscape + KEGG >> << Bioconductor >> Grading: Homework Assignments/quizzes 20% Exam 1 10% Exam 2 10% Exam 3 10% Software projects 25% Final Examination 25% NOTE: homework assignments concern typical problems and software practice. Course Outline: PART 1: Structure and properties of different biomolecules organelles A. Amino-acids and protein; Protein domains, post-translational modification; Protein trafficking, Protein degradation and turnover B. Nucleic acids C. Lipids, membranes and transport D. Carbohydrates E. Mitochondrial function PARTII: Signal transductions and metabolism with clinical correlations A. Cofactors and vitamins B. Fundamentals of signal transduction C. Bioenergetics and Oxidative metabolism D. Carbohydrate metabolism E. Lipid metabolism F. Amino acid metabolism G. Nucleic acids metabolism PART III: Biochemistry of hormones and nutrients A. Hormones and hormonal cascades B. Peptide hormones and amino-acid derived hormones and signaling C. Steroid hormones and signaling D. Basic nutritional constituents E. Clinical correlations PART IV: Gene regulation, epigenetics and human disease A. Chromatin remodeling and transcription through chromatin B. Histone modification and histone code hypothesis C. Epigenetic mechanism of gene activation and silencing Prerequisites: Biochemistry, Calculus II Molecular Control of Metabolism and Metabolic Disease: Examination of various physiological states and how they affect metabolic pathways. Discussion of a number of special topics related to the unique roles of various tissues and to metabolic pathways in disease states, including adipocyte biology, beta-cell biology, epigenetics, inflammation, and aging related diseases. Goals: -Understand the adjustments in fuel utilization and in the regulation of metabolic pathways required by mammalian fast-feed cycles. -Examine how various physiological states affect metabolic pathways. -Discuss the unique roles of various tissues and metabolic pathways in disease states, including diabetes, cancer, inflammation, and age-related disease processes. -Synthesize knowledge and use insight to better understand the molecular control of metabolism and metabolic disease. Grade: Course Exercises 10% Quizzes 15% Software projects 45% 2-3 Exams 30% Prototypical text: Textbook of Biochemistry with Clinical Correlations, Thomas M. Devlin BIOCHEMICAL SOFTWARE --> Concerning particular topics, bonds and biochemical processes related to metabolism. Will have extensive observation and description of properties, characteristics of the processes, mechanisms, outputs, etc. in terms of organic chemistry and biochemistry. Software activity goes along with the respective report. There will be numerous cases given to students (in groups and individually). MUST connect with predominant or modern research, hence some references from journal articles, etc. --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> ESSENTIALS --> << COPASI, Pathvisio + Cytoscape + KEGG >> << Micro-Manager Open Source Microscopy software with ImageJ, JoVE (video journals), UIUC-Virtual Microscope >> << Bioconductor >> SIMULATION LABS --> Conventional molecular simulations play a significant role in understanding the molecular mechanisms underlying metabolic diseases. These simulations provide insights into the interactions between biological molecules, their dynamics, and structural changes. Applications of conventional molecular simulations in the context of metabolic diseases that will be pursued (where order may change to suit course progression): 1. Carbohydrate Metabolism & Carbohydrate-response element-binding protein. 2. ChREBP tasks 3.Drug Discovery and Development -- Application: Molecular simulations are used to model the interactions between small molecules (drugs) and target proteins involved in metabolic pathways. This aids in predicting binding affinities, understanding drug-receptor interactions, and optimizing drug candidates for the treatment of metabolic diseases such as diabetes and obesity. 4.Protein-Ligand Interactions -- Application: Simulations can elucidate the dynamic behavior of enzyme-substrate or receptor-ligand interactions involved in metabolic processes. This information is crucial for understanding the molecular basis of metabolic diseases and designing targeted therapies. 5.Protein Conformational Changes -- Application: Metabolic diseases are often associated with changes in protein conformations. Molecular dynamics simulations can provide insights into how proteins change their shapes over time, helping to understand the structural basis of diseases and identify potential therapeutic targets. 6.Lipid-Protein Interactions -- Application: Simulations can explore the interactions between lipids and proteins, particularly in the context of diseases like atherosclerosis. Understanding lipid-protein interactions helps in studying the formation of lipid plaques and developing strategies to prevent or treat cardiovascular diseases. 7.Metabolite Binding and Recognition -- Application: Molecular simulations can be employed to study how metabolites interact with proteins, receptors, or enzymes. This is relevant for understanding the regulatory mechanisms in metabolic pathways and identifying potential targets for intervention. 8.Protein Folding and Misfolding -- Application: Simulations can shed light on the folding pathways of proteins involved in metabolic processes. Aberrant protein folding and misfolding can lead to diseases like amyloidosis, and simulations help in understanding these processes at the molecular level. Transport Processes -- Application: Simulations can be used to study the transport of molecules across biological membranes, such as glucose transport in diabetes. Understanding the dynamics of transporters aids in developing strategies to regulate metabolic processes. 9.Enzyme Catalysis-- Application: Molecular simulations provide insights into the catalytic mechanisms of enzymes involved in metabolic pathways. Understanding enzyme kinetics and reaction mechanisms is crucial for developing interventions in metabolic diseases. 10.Structural Dynamics of Cellular Components -- Application: Simulations can explore the dynamics of cellular components, including organelles and membranes, providing a comprehensive view of how cellular structures contribute to metabolic processes and how disruptions can lead to diseases. COURSE OUTLINE --> A. INTERMEDIARY METABOLISM --Course Introduction & Carbohydrate Metabolism --Carbohydrate Metabolism --Carbohydrate Metabolism & Carbohydrate-response element-binding protein B. ChREBP --Fatty Acid to Glucose? Pyruvate Metabolism, Steady-States --Ketone Body Metabolism & b-oxidation --TCA Cycle & Carbonyl Chemistry --Glycogen Metabolism & Gluconeogenesis (Anderson/Rhoads) --Lipogenesis & Lipoprotein Metabolism --Cholesterol Metabolism C. MITOCHONDRAL METABOLISM --Mitochondrial metabolism --Mitochondrial metabolism --Mitochondrial metabolism D. METABOLIC FLEXIBILITY --The Unfolded Protein Response & Autophagy --Cycles, shuttles, and shunts --Metabolic signaling; primary & secondary messengers --GL/FFA cycle, hormonal regulation of lipolysis, lipid droplet biology --b-cell biology and diabetes E. SIGNALLING AND REGULATION --Insulin signaling & insulin resistance --mTor & Regulatory Nodes --Cold exposure and sympathetic nervous system metabolism --Cold exposure and sympathetic nervous system metabolism --Cancer Metabolism Augmented with the following: Ralph J. DeBerardinis, Navdeep S. Chandel, Fundamentals of Cancer Metabolism.Sci. Adv.2,e1600200(2016). Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016 Jan 12;23(1):27-47 F. INTEGRATED METABOLISM --Exercise, aging, & metabolic disease --Hypothalamic control of metabolism and circadian rhythms --Epigenetics --Alzheimer’s and other degenerative diseases --Inflammation Prerequisites: Metabolism, Upper Level Standing Bioprocess Engineering: Introduction to biochemical and microbiological applications to commercial and engineering processes, including industrial fermentation, enzymology, ultrafiltration, food and pharmaceutical processing and resulting waste treatment. Enzyme kinetics, cell growth, energetics and mass transfer. The objective of the course is to introduce fundamental bioprocess engineering concepts often reserved for chemical engineers. Knowledge, resources and economics are seemingly perpetual factors that drive success. The emphasis will be application of the following core chemical engineering concepts to biological problems -- A. Material and heat balances-distributed throughout B. Reaction kinetics and reactor design Enzyme kinetics, fermentation kinetics, batch/fed-batch/continuous bioreactor, recycle, bioreactors-in-series, heterogeneous catalysis, biological wastewater treatment C. Transport Immobilized enzyme/cell reactor, biofilm reactor, mixing, oxygen mass transfer, sterilization, separation and product purification D. Thermodynamics Energy efficiency, yield E. System dynamics Steady-states, Metabolic network, bioreactor stability, mixed culture F. Applied mathematics - distributed throughout G. << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> H. Use of Process Simulators ASPEN (or cheaper substitute) BEST: Biochemical Engineering Simulation Technology. National Renewable Energy Laboratory 1996. NREL/MP-425-20548. Task Number IP443431 Use of Process Simulators COCO (+ ChemSep) or DWSIM (+ ChemSep) or ESMO Note: conventional lectures supplemented by in-class demonstrations of computer simulations and computer simulations assignments; possibly software such as ASPEN or COCO (+ ChemSep) or DWSIM (+ ChemSep) or ESMO will have relevance. Such software will help students gain a better feel or sense of meaningfulness/purpose for the topics, various models and operating parameters. Knowledge of R + RStudio will be beneficial. Expected primitives acquired from prerequisites to accompany A through G prior throughout course -- 1.Knowledge of relevant ODEs and general solutions 2.Ability to solve simple ODEs using explicit numerical methods 3.Working knowledge of RStudio along with a working knowledge of Excel. For R will likely incorporate biological & ecological packages if constructive 4.Ability to solve sets of linear and nonlinear algebraic equations numerically via CAS such as R + RStudio 5.Familiarity with matrix multiplication structure Priorities are not the whosoever in a mathematics department Ability with 2 by 2 matrices shows you know what you’re doing M by N matrices concern graphing calculators and R + RStudio Professionals don’t care about pig pen finesse on a board, but rather the purpose of the models; understand your priorities and real talent/ability. Course Text: Bioprocess Engineering, Basic Concepts, Michael L. Shuler and Fikret Kargi, Prentice Hall, 2001 Assisting Text (whenever deemed practical and malleable): John Villadsen, Jens Nielsen, and Gunnar Lidén. Bioreaction Engineering Principles, Third Edition, Springer Publishing, 2011. We will touch upon the issues of the balance between biotechnology's benefit to the society and profiteering, and the regulatory issues. Course Assessment: Homework 10% Computer simulation assignments 35% Structured and administered to construct and implement practical and sustainable simulations and crucial observations often encountered in Bioprocess Engineering Immersion and Retention Activities Labs 3 Midterm exams 30% Term Project 25% Note: labs with software tools use will accompany course topics extensively. Will develop simulation processes/projects relevant to topics. Each simulation project to accompany a specific set of course topics. Course Outline: -Biochemical & Bioprocess Engineering (Ch 1) -Biology & Biochemistry (Ch 2) -Enzyme Kinetics & Immobilization (Ch 3) -Genetics & Cellular Control Systems (Ch 4) -Genetic Engineering (Ch 8, 14) -Metabolism (Ch 5) -Stoichiometry (Ch 7) -Cell Growth Kinetics (Ch 6) -Bioreactor Design & Operation (Ch 9) -Scale-up, Heat/Mass Transfer, Instrumentation, Control (Ch 10) -Product Purification & Recovery (Ch 11) -Mixed Culture (Ch 16) Prerequisites: Genetic Engineering & Technology, Organic Chemistry I, Biochemistry, ODE, Upper Level Standing. Bioprocess Engineering Lab: Goals of Course --> 1. Understand the experimental and mathematical frameworks underlying the growth of biological organisms and the production of macro and small molecule products. 2. Analyse reaction stoichiometry for biochemical processes. 3. Evaluate different methods for producing biological molecules, including cell culture and protein expression systems. 4. Develop strategies for metabolic engineering of biological organisms, to manufacture useful chemicals. 5. Select and sequence purification processes for biological products. 6. Design and evaluate drug delivery pathways and tissue engineering methods. 7. Understand protein and cellular engineering approaches. Software Tools --> R + RStudio Excel << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> Use of Process Simulators ASPEN (or cheaper substitute) BEST: Biochemical Engineering Simulation Technology. National Renewable Energy Laboratory 1996. NREL/MP-425-20548. Task Number IP443431 COCO (+ ChemSep) or DWSIM (+ ChemSep) or ESMO Note: conventional lectures supplemented by in-class demonstrations of computer simulations and computer simulations assignments; possibly software such as ASPEN or COCO (+ ChemSep) or DWSIM (+ ChemSep) or ESMO will have relevance. Such software will help students gain a better feel or sense of meaningfulness/purpose for the topics, various models and operating parameters. Knowledge of R + RStudio will be beneficial. Expected primitives acquired from prerequisites to accompany A through G prior throughout course -- 1.Knowledge of relevant ODEs and general solutions 2.Ability to solve simple ODEs using explicit numerical methods 3.Working knowledge of RStudio along with a working knowledge of Excel. For R will likely incorporate biological & ecological packages if constructive 4.Ability to solve sets of linear and nonlinear algebraic equations numerically via CAS such as R + RStudio 5.Familiarity with matrix multiplication structure Priorties are not the whosoever in a mathematics department Ability with 2 by 2 matrices shows you know what you’re doing M by N matrices concern graphing calculators and R + RStudio Professionals don’t care about pig pen finesse on a board, but rather the purpose of the models; understand your priorites and real talent/ability. Course Text: TBD. Additionally, literature from prerequisite can assist well. Syllabus --> ---Introduction to Biochemical Engineering Historical background, Interdisciplinary approach, Integrated Bioprocess systems, Unit Operations in Bio-processes. ---Microbial Growth Kinetics Batch Culture Continuous Culture –Multistage systems, Feedback systems Fed Batch Culture –Variable volume, fixed volume, Cyclic. Applications. ---Design of Fermentor Introduction, Basic Functions, Body construction, Aeration and Agitation, Maintenance of aseptic conditions, Control of parameters, Valves and steam traps, Variants of fermentation vessels. ---Aeration and Agitation Introduction, Oxygen requirement in fermentations, Oxygen supply, Determination of K-La values, Fluid rheology, Factors affecting K-La values, Balance between oxygen demand and supply, Scale up and Scale down. ---Basic Outline of fermentation process and purification of fermentation products Introduction, Range of fermentation process, Components of fermentation process, Disruption of cells, Precipitation, Filtration, Centrifugation, Liquid-Liquid Extraction, Chromatography, Membrane processes, Drying, Crystallization. Laboratory Experiments --> Note: software tools use will accompany lab experiments extensively as precursor or pre-labs; assumed high knowledge and skills from prerequisite. Will develop (advance and extensive) simulation processes/projects relevant to labs. Each advance and extensive simulation project to accompany a specific set of labs. 1. Determination of Oxygen Transfer rate 2. Determination of K-La value 3. To obtain growth curve of bacteria under batch culture 4. To obtain growth curve of bacteria under fed batch culture 5. To carry out precipitation of protein 6. To perform column chromatography 7. To perform drying operation. 8. To perform crystallization operation Prerequisites: Bioprocess Engineering Drug Metabolism: The proposed course in Drug Metabolism is designed to provide the students an understanding of (i) the key role played by metabolic processes in the design and development of safe and efficacious therapeutic agents (ii) the relevant metabolic transformations and the enzymes (enzymology) responsible for these transformations (iii) the contemporary techniques in cell and molecular biology, chromatography, mass spectrometry, and spectroscopy used to study metabolic processes and the role of specific metabolic enzymes in the metabolism of drug molecules. Course to be instructed by industry professionals (scientists from pharmaceutical companies and faculty). Texts & Notes: TBA Tools --> << COPASI, Pathvisio + Cytoscape + KEGG >> << Bioconductor >> Assessment --> Group Assignments: 20% 4 Development phases 0.4 Report 0.5 Presentation 0.1 Lab: 20% 4 Exams: 60% Lab --> PART A OPTIONS The following articles will be applied to develop lab experimentation. It’s important that they are harmonic with lecturing Farrell, S and Hesketh, R. (2002). An Introduction to Drug Delivery for Chemical Engineers. Chemical Engineering Education http://users.rowan.edu/~hesketh/hesketh/cee%20drug%20delivery.pdf Farrell, S., Savelski, M., Hesketh, R. and Slater, C. S. (2006). Experiments in Drug Delivery for Undergraduate Engineering Students. American Society for Engineering Education Farrell, S. and Vernengo, J. (2012). A Controlled Drug-Delivery Experiment Using Alginate Beads. Chemical Engineering Education, v46 n2 p97-109 Farrell, S. et al (2013). An Experiment to Introduce pH-responsive Hydrogels for Controlled Drug Delivery: Mechanical Testing. 120th ASEE Annual Conference and Exposition Puperi, D. (2019). Extending an Alginate Drug Delivery Experiment to Teach Computational Modelling and Engineering Analysis to 1st Year Biomedical Engineering Students. Paper Presented at 2018 ASEE Gult-Southwest Section Annual Meeting, AT&T Executive Education and Conference Center, Austin, TX 78705 < https://peer.asee.org/31598 > Ranjan, A. and Jha, P.K. (2020). Experiments and Modelling of Controlled Release Behaviour of Commercial and Model Polymer-Drug Formulations using Dialysis Membrane Method. Drug Deliv. and Transl. Res. 10, 515–528. Werzer, O. et al (2019). Drug Release from Thin Films Encapsulated by a Temperature-Responsive Hydrogel. Soft Matter, 15, 1853-1859 PART B OPTIONS Note: for the following two subjects in particular, after analysis to pursue modelling and simulation of the delivery processes with software encountered in the biochemistry course and metabolism course. Vilar G, Tulla-Puche J, Albericio F. (2012). Polymers and Drug Delivery Systems. Curr Drug Deliv. 9(4): 367-94 Drug Delvery by Supramolecular Sesign Group Assignments --> For known authorized drugs in a given market, after medical profiling, student groups will pursue modelling and simulation of the delivery processes with software encountered in the biochemistry course and metabolism course. Includes ideal environments for optimal function, mechanisms, functional groups, activation sites, by-products, etc., etc., etc. Topics --> LECTURE 1 Introduction to Drug metabolism --Historical Perspective and General Principles --Drug metabolism as a mechanism for clearance of therapeutic agents; pharmacokinetic concepts, metabolic clearance, its role in the total clearance of drug molecules, Stereochemistry in Drug Metabolism general concepts, role in drug metabolism and pharmacokinetics, regulatory issues. Role of drug metabolism in the design and development of safe and efficacious therapeutic agents, regulatory issues. LECTURES 2-5 Chemistry of Metabolic Reactions --Oxidation --Reduction --Hydrolysis --Conjugation LECTURE 6 Biochemistry of Cytochrome P450 --Classification --Isozymes --Multiplicity and Substrate Specificity --Localisation --Variability LECTURES 7-8 Induction of Drug Metabolizing Enzymes --Mechanisms --in vitro models to asses induction --Clinical considerations/implications LECTURE 9 Intestinal Oxidative Metabolism --Intestinal cytochrome P450 enzymes --Role of intestinal cytochrome P450 enzymes in drug-drug interactions LECTURE 10 Phase I (non-P450) Enzymes --Oxidative --Reductive --Hydrolytic LECTURE 11 Inhibition of Drug Metabolizing Enzymes --Mechanisms --In vitro assessment --Clinical considerations/implications LECTURE 12 Phase II Enzymes --Introduction --Glutathione Transferases --Reaction and substrates --Physiological considerations --Expression and Regulation --Species and strain differences LECTURE 13 Glucuronosyl transferases and sulfotransferases: --Classification --Reactions and classes of substrates --Physiological considerations and cofactors --Expression and regulation --Species and strain differences LECTURE 14 In Vitro Model Systems for Transport --In vitro techniques to study drug transport --Transport models to elucidate and predict transporter-based drug interactions LECTURES 15-16 Hepatobiliary and Renal Disposition – Hepatic and Renal Transport --First Pass Effect --Mechanisms --Hepatic and renal clearance --Classes of drugs excreted --Pharmacological factors influencing biliary excretion of xenobiotics: methods for examining biliary and renal excretion enterohepatic recirculation and actors that influence enterohepatic cycling. --Transporter-based hepatic and renal toxicity LECTURE 17 Intestinal Transport and Transporters --Intestinal transport mechanisms --Absorptive and Barrier properties of intestinal epithelium --Absorptive and Secretory transporters --Uptake and efflux transporters --Transporter-based drug-drug interactions LECTURE 18 -20 Analytical Techniques for Studying Drug Metabolism --NMR --LC/MS --Stable isotopes --Radioisotopes LECTURES 21-22 Metabolism-based Drug Toxicity --Mechanisms of toxicity - reactive metabolites --Genotoxicity --In vitro systems to assess toxicity --Drug- and metabolite-induced hepatotoxicity --Implications in drug discovery/development LECTURES 23-25 Pharmacogenomics of Drug Metabolizing Enzymes and Transporters --Basic Concepts --Pharmacogenomics of Drug Metabolizing Enzymes and Transporters --Case studies and clinical implications --Regulatory impact LECTURE 26. Integration of Drug Metabolism/pharmacokinetic Function in Drug --Discovery and Development --Review of the DM/PK function within pharmaceutical R & D --DM/PK studies in drug discovery and development --Case histories LECTURE 27. Review and Key Learnings from the course Prerequisites: Organic Chemistry I, Biochemistry, Metabolism, Upper level Standing Protein Engineering: Course covers the principles, techniques, and applications of designing and modifying proteins for various purposes. Applications of interest include (but not limited to): vaccines, therapeutics, diagnostics, drugs, material science, industrial processes. Course outline dictates the types of applications and labs. Each lab will include software development as a precursor or pre-lab to the detailed labs throughout the term. Emphasis on reinforcing skills with software: << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> << Bioinformatics tools. Note: could be R based >> ASSESSMENT --> Lab Quizzes 25% Labs Operations 50% Attendance Punctuality Behaviour Content/Substance Student Groups Projects 25% Attendance Punctuality Behaviour Content/Substance Presentations COURSE OUTLINE --> WEEK 1: Introduction to Protein Engineering Overview: Introduction to the field of protein engineering, historical context, and key principles. Topics: Importance of protein engineering, applications in biotechnology and medicine. WEEK 2: Protein Structure and Function Lectures: Review of protein structure and function. Lab Activity: Use bioinformatics tools to analyze protein structures and predict functional domains. WEEK 3-4: Rational Protein Design Lectures: Principles of rational protein design. Lab Activity: Design a protein variant using rational design principles and modeling tools. Prerequisites: WEEK 5-6: Directed Evolution Lectures: Overview of directed evolution techniques. Lab Activity: Perform a directed evolution experiment using a model protein target. WEEK 7-8: Enzyme Engineering Lectures: Engineering enzymes for enhanced catalytic activity and specificity. Lab Activity: Design and optimize an enzyme through directed evolution. WEEK 9-10: Antibody Engineering Lectures: Engineering antibodies for therapeutic applications. Lab Activity: Design antibody variants with improved affinity or altered effector functions. WEEK 11-12: Protein Expression and Purification Lectures: Strategies for efficient protein expression and purification. Lab Activity: Express and purify engineered proteins using different expression systems. WEEK 13: Computational Approaches in Protein Engineering Lectures: Introduction to computational tools for protein engineering. Lab Activity: Use computational methods to predict protein stability, binding sites, and design variants. WEEK 14-15: Protein-Protein Interaction Engineering Lectures: Strategies for modifying and engineering protein-protein interactions. Lab Activity: Design and analyze protein variants to alter or enhance protein-protein interactions. WEEK 16-18: Student Groups Research Projects Project Assignment: Students design and propose a protein engineering research project. Proposal Presentation: Students present their project proposals to the class for feedback. Lab Activities: Students execute their protein engineering research projects. Progress Reports: Periodic progress reports and discussions. Presentations: Students present their research projects to the class. Prerequisites: Calculus II, ODE, General Physics I, Organic Chemistry I, Genetic Engineering & Technology, Molecular Biology II, Senior Standing.
Note: augmenting curriculum of biological and biochemical sciences with the critical hands on research areas, starting from upper level sophomore students for “winter and “summer” semesters. Likely to be collaboration among constituents (of free will) under biology. FOR ACTIVITIES IN THE “SUMMER” AND WINTER” SESSIONS ALL PARTICIPATING STUDENTS, ASSISTING/ADVISING INSTRUCTORS AND PROFESSORS MUST BE OFFICIALLY RECOGNISED; REQUIRES BOTH CIVILIAN ID AND STUDENT/FACULTY ID FOR CONFIRMATION OF INDIVIDUAL. THERE WILL ALSO BE USE OF IDENTIFICATIONS FOR ACTIVITIES FOR RESPECTIVE SESSION. SECURITY AND NON-PARTICIPATING ADMINISTRATION WILL ONLY IDENTIFY RESPECTIVE ACTIVITY BY IDENTIFICATION CODE. SECURITY AND NON-PARTICIPATING ADMINISTRATION MUST NEVER KNOW WHAT ACTIVITIES IDENTIFICATION CODES IDENTIFY: < Alpha, Alpha, Alpha, Alpha > - < # # # # # > - < session > - < yyyy > Such biological sciences activities will also warrant criminal background check (CBC) in order to participate. Severely threshold may vary depending on administration. Administrators will provide dated letters of confirmation of thorough CBC to student affairs and other appropriate administration. Such may also include screening that’s parallel to customs & immigration processing where certain levels of criminal history warrants rejection. Email and physical letters with data. Such CBC protocol will not explicitly identify any particular titles or descriptions of any activity, rather, will only convey code as above. Innovation and technology being key, projects may not be confined to the mentioned two curriculum areas, if they are constructive, practical, economic, and appealing to progressive and developed society. For projects establish control (group, specimen, samples, etc.) whenever relevant. There will be a secure database archive for all participants and supervision constituents for respective activity chronologized. Activities will be classified. VARIOUS SOFTWARE AVAILABLE It may be the case some activities can be grouped and given a major title together; however, detailed descriptions will be required. Activities repeated can be added to transcripts upon successful completion. Repeated activities later on can be given a designation such as Advance “Name” I, Advance “Name” II. As well, particular repeated activities serve to towards developing true comprehension, competency and professionalism. Any media developed is not geared to pop culture and minority trends or stereotypes. Activities will be field classified. Particular activities of interest being stationary: 1. Transduction Lab Orchestrating the process and observing the results, namely the effectiveness or the intended quality or characteristic pursued in a bacterial or mammalian cell. Useful by-products would be cnstructive. Keep economic as much as possible. 2. Bacterial Conjugation reinforcement https://www.sas.upenn.edu/LabManuals/biol275/Table_of_Contents_files/16-BacterialConjugation.pdf Will incorporate competent use of microbial, viral, pathogen and EPA software from software portfolio. Can be compared with curve fitting of data, and regression modelling can also be done. 3. Polymerase Chain Reaction and common tactics involving the process (analysis and experimentation) -Operational Theory & Logistics and exponential amplification -Parameters that affect PCR -Denaturation -Annealing of primers to template DNA -Number of cycles -Experimentation: http://www2.southeastern.edu/Academics/Faculty/jtemple/486/experiment%203.pdf What role do each of the following play in PCR - dNTPs, taq polymerase, primers? If temperature rises above 94 C there’s chance of water evaporation. How is it prevented? What is the use of buffer in the reaction mixture? Police investigates a homicide. From crime scene they got hair samples and semen from the body. What could be done to identify the murderer? Note: interested in other types of DNA for PCR experimentation as well. Strawberry DNA as one example is easy to extract. -Simulation examples: Animated Tutorial 9.5 Polymerase Chain Reaction Simulation Fellermann, H. et al, The PCR Simulator: An On-line Application for Teaching Design of Experiments and the Polymerase Chain Reaction, bioRxiv 415042. Concerns a run through then testing of understanding with simulator. Succeed with actual processing regardless. -Actual lab processing version of simulation above to be done. 4. Cellular Signalling Svoboda, K. K., & Reenstra, W. R. (2002). Approaches to Studying Cellular Signaling: a Primer for Morphologists. The Anatomical record, 269(2), 123–139. 5. CRISPR A revolutionary new technology for genome editing that provides unheard-of ease, efficiency, and low cost. It’s now universal use in laboratories worldwide has led to subsequent breakthroughs in inherited disease, HIV, malaria, retinitis, and cancer. Pursuit towards adopting the technology for in-house experimentation. To focus on applications of CRISPR technology as a platform for [genome editing] and functional genomics. The activity will consist of lectures from experts in the field and a hands-on laboratory experience demonstrating CRISPR editing both in vitro and in vivo. Expert professional leadership will address topics in genome editing and CRISPR-Cas9 research, including basic and enhanced CRISPR methods, cellular repair mechanisms, regulation of gene expression, bioinformatics, applications to various organisms, and bioethics. TBD, will include: All protocols for lab works, rubric for grading lab report, major CRISPR publications, news articles and popular science CRISPR articles. Lap reports and participation with integrity are crucial towards any measure of real accomplishment. Constitution to advantage of biochemistry, cellular biology, molecular biology and microbiology software and tools mentioned. Pursue CRISPR design tools as well. -Acquisition Assay (operations sequence) Instruction: Welcome, Course Introduction, Lab Safety In Lab: Transformation of Cas1 and Casa2 plasmids, plating of bacteria, take initial sample Instruction: CRISPR immunity In Lab: Take additional samples, PCR of CRISPR locus, run agarose gel, pick colonies for overnight cultures Instruction: Structure and Function of Cas9 In Lab: miniprep cultures and send for sequencing Instruction: Genome Editing and DNA repair In Lab: Analyse sequencing results and present. -In Vitro Cleavage Assay Instruction: gRNA Design In Lab: PCR, Run agarose gel, Set up ivt overnight Instruction: Reducing Off-Target Effects In Lab: Purification, Run RNA gel to check product Instructor: CRISPR Application - Plants In Lab: in vitro cleavage assay Instruction: CRISPR Applications: Human Therapeutics In Lab: Presentation of results. Bioinformatics Practical - gRNA design -In Vivo Editing Instruction: CRISPR Applications: Bioenergy In Lab: Transformation of control RFP expression plasmid, Transformation of Cas9 gRNA RFP editing plasmid, Plate cells. Instruction: CRISPR Applications: Model Systems In Lab: Observe results, pick colonies, inoculate overnight cultures with and without selection. Instruction: CRISPR Ethics/ Policy In Lab: Plate cultures, Ethics discussion Instruction: CRISPR Ethics/ Policy In Lab: Observe results. Presentations of final project. The next phase will be designing and implementing experiments(s) to critique or validate the following: Gebre, M., Nomburg, J., L., and Gerwurz, B., E., CRISPR-Cas9 Genetic Analysis of Virus-Host Interactions, Viruses 2018 Jan 30; 10(2) Applying PCR to amplify CRISPR arrays and analyse spacer content for specifically targeted CRISPRs and for organisms with sufficient representation in public databases to design reliable PCR primers. 6. Immunoproteomics & Immunochemistry --Host and Viral Determination of Infection Outcome Consider common place viruses. How does the interaction between host and virus influence infection outcome and disease progression? What are the genetic immunological signatures of an effective host immune response against such viruses? Utilizing samples from well characterized cohorts. Samples to be assayed via sequencing, single-cell technologies and cellular immunology tests. Concerns means to inform vaccine design and future immune-therapy. --Design and implement experiment(s) to critique or validate the following: Falisse-Poirrier, N. et al, Advances in Immunoproteomics for Serological Characterization of Microbial Antigens, Journal of Microbiological Methods 67 (2006) 593–596 --For the following link provided design and implement experiments(s) of the model described for broad range detection of various viruses or bacteria (instead of Helicobacter Pylori): Haas, G., et al, Immunoproteomics of Helicobacter Pylori Infection and Relation to Gastric Disease, Proteomics 2002, 2, 313–324 --Design and implement experiment(s) to critique or validate the following: Israr, B., Kim, J., Anam, S., and Anjum, F., R., Lactic Acid Bacteria as Vectors: A Novel Approach for Mucosal Vaccine Delivery, J Clin Cell Immunol 2018, 9:2 --Design and implement experiment(s) to critique or validate the following: Shonyela, S., M. et al, New Progress Regarding the Use of Lactic Acid Bacteria as Live Delivery Vectors, Treatment of Diseases and Induction of Immune Responses in Different Host Species Focusing on Lactobacillus Species, J Prob Health 2017, 5:4 7. Biochemistry of anaesthetics and antiseptics Note: for the case of antiseptics treatment, treat accordingly for natural and synthesizingsources, and the associated automatic control. Note: much interest in the metabolic pathways Note: biochemical software will accompany development Modelling Simulations, reactions, chemical characteristics ---Phase 1 Comprehension of the terms hypoalgesia and hyperalgesia Comprehension of the biological automatic control regulation relevant involving organs, nociceptors, enzymes and pathways concerning involving anaesthetics/analgesics, endorphins, enkephalins, and dynorphins. Processes of consideration: Transmission Upwards (thalamus & distribution, reticular formation, amygdala) Referred Pain Gate theory Phantom Pain Identification of nociceptor types: thermal nociceptors, mechanical nociceptors, chemical nociceptors, polymodal nociceptors, silent (or sleeping). Identify in nociceptors the constituents or unique design that gives the special function. When there is significant damage to tissue, several chemicals are released into the area around the nociceptors. This develops into what is called the "inflammatory soup," an acidic mixture that stimulates and sensitizes the nociceptors into a state called hyperalgesia. Some of the chemicals involved: “Inflammatory soup” (biochemical mechanism, by-products and simulation) Prostaglandins released by damaged cells (biochemical mechanism, by-products and simulation) Potassium is released by damaged cells (biochemical mechanism, by-products and simulation) Serotonin is released by the blood platelets (biochemical mechanism, by-products and simulation) Bradykinin is released by blood plasma (biochemical mechanism, by-products and simulation) Histamine is released by mast cells (biochemical mechanism, by-products and simulation) “Substance P” (biochemical mechanism, by-products and simulation) Histamine (biochemical mechanism, by-products and simulation) Antihistamines and reaction with histamine (biochemical mechanism, by-products and simulation) Marijuana will be treated concerning point, region, conditions, etc. for beginning influence in the regulatory/nervous/endocrine system. Includes biochemical mechanism, by-products and simulation. There are tissues that contain nociceptors which do not lead to pain. In the lungs, for example, there are "pain receptors" which cause you to cough, but do not cause you to feel pain. Establish the biochemical process and operation in the regulatory/nervous/endocrine system that permits coughing rather than pain. There are some people born with a genetic inability to feel pain. It’s quite rare. Identify genes associated to pain stimulus. What is the genetic structuring for the inability to have the have stimulus mechanism? What isn’t being produced? NOTE: for recognition of substances various spectroscopy types will be employed and compared with databases. Extraction of anaesthetics/analgesics from saliva and other body areas. In laboratory environment, to observe how such anaesthetics/analgesics react with the following chemicals associated to pain (rate and by-products). Extraction of endorphins, enkephalins, and dynorphins if possible. Software provided to be used as simulation prediction, whereas lab activity to validate with other empirical observations. An elaborate and economic scheme for observation to be developed and administered; possibly includes rate of reaction, by-products (quantities if feasible). Reaction analysis (rate of reaction, by-products) of such opioids to be compared with extractions from saliva, as well to industrially synthesized treatments like aspirin, naproxen sodium, acetaminophen, ibuprofen, Vicodin (if feasible), oxycodone (if feasible), codeine (if feasible), and others. Automatic control descriptions for each; heroine is also possible. Then the reaction analysis of such natural and artificial substances can be compared with natural herbs such as chamomile, willow bark, turmeric, cloves, ginger (pain & inflammation), etc. Based on automatic control of pathways determine optimal means of administering herbal remedies and synthetics, respectively. Note; for herbal remedies the uniqueness of one particular herb “bio product” its optimal administering may or may not be the same as others; same goes for synthetics. Biochemical response with melatonin, adrenaline and dopamine. Establish definitively the respective function difference for such mentioned three unique to opioids, anaesthetics/analgesics. Concerned with exercise, fear & shock, aggression, and diabetes which includes automatic control descriptions for pathways. If diabetes is to be differentiated then, so be it. For melatonin, adrenaline and dopamine, respectively, identify the concerning point, region, conditions, etc. for beginning influence in the regulatory/nervous/endocrine system; concerned with respective biochemical mechanism and simulation as well. Extraction of melatonin, adrenaline and dopamine from the most feasible places. In laboratory environment, to observe how such anaesthetics/analgesics react with the following chemicals associated to chemicals in respective process (with by-products). NOTE: Trials and/or samples for the various laboratory activities may be a necessity, and also dependent on economics (of time, money, resources, utilities, etc.). ---Phase 2 Like phase 1 study and lab activities saliva will be also used for extraction of antiseptics. However, before such, develop an analogous sequence of operations top-down observed in phase 1; likely the nervous system, adrenaline, melatonin, dopamine and such will not factor in (as much). Figure all that out. Mechanisms and processes may only remain at regions of concern at the cellular level. Saliva constituents of concern: referencing Wikipedia’s page “Wound Licking” under “Mechanism”. Other pursued interests for antimicrobial substances with experiments and methods: Fruits Sorrel, balsam of Peru, and other cultural plants/trees Iodine, hydrogen peroxide, various alcohols, phenol, Dakin’s solution, Different antibiotics Octenidine dihydrochloride Polyhexamethylene biguanide, PHMB Super oxidized solutions NOTE: for recognition of substances spectroscopy types or chemical reactions will be employed and compared with databases. Synthesis will be carried out for acquisitions. In petri dishes and what not microbial organisms to be grown coming from various sources such as river water, wastewater, dirt, bathroom surfaces, decomposing sustenance, general surfaces, etc. NOTE: Trials and/or samples for the various laboratory activities may be a necessity, and also dependent on economics (of time, money, resources, utilities, etc.). Microbial constituents (bacterial, viral or other) should be identified. Identify place in the Phylogenetic tree and genus; analogy for viruses and other microorganisms identified. Growth rates should be identified (theoretical versus actual) for respective type of bacteria, virus, etc. Software to accomodate mathematial modelling. Treatment of evolved resistance (on both biochemical and genetic level). 8. Programmable bacterial (or algae) for the bio-synthesis of compounds of interest such as insulin (or whatever interests) Laboratory production of [insulin] by recombinant DNA technology with bacteria/algae population growth. Fermentation or whatever process to be coerced in a isolated environment; characteistics of environment must be particular and stable to acquire both optimal bacteria/algae growth and optimal insulin production. Satefy protocols and regulation with microbes Review of recombinant DNA technology Process and logistics for yeast with recombinant DNA technlogy Environmental conditions regulation For optimal bacteria/algae growth & optimal insulin production Identify & comprehend models, modelling, parameters & measures Biochemistry and Organic Chemistry modelling Analysis with possible by-products Stoichiometry Growth models based on whatever parameters prefrences Production expectation at time Tfinal Tools and structure for data gathering and assurance Description and profiling of the biprocessing system Means of [insulin] separation & gathering for storage & long term stablity Walk-through of operation procedures of bioprocessing system With logistical saftey protocols for whatever phases With tools and structure for data gathering and assurance Review of preparation for the relevant phases in development Implementation of bioprocessing Means of confirming [insulin] product Means of determining gross production and if consistent with preliminary modelling expectations Economics and innovation with technologies Software of interest R + R Studio Excel COCO (+ ChemSep), DWSIM (+ ChemSep) << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> A possble assist: 9. Competence and economics in Viral and Bacterial detection --Symptomology of specified common viruses. --Diagnosis can be done by direct detection of a virus in clinical specimens or by detecting the presence of Ab's to the virus in serum (acute infection is diagnosed by the presence of IgM antibody or by a 4X rise in IgG titer between acute and convalescent sera). Will be done in labs. Methods for direct virus detection: i). Isolation in cell culture ii). Electron microscopic examination of specimens iii). Immunofluorescence staining of specimens and microscopic examination (presence of antigen) iv). Enzyme immunoassay (presence of antigen) v). Polymerase chain reaction (presence of nucleic acid). Possible guide, but may choose variation and targets depending on resources: Adjou Moumouni, P. F et al. (2015). Molecular detection and characterization of Babesia bovis, Babesia bigemina, Theileria species and Anaplasma marginale isolated from cattle in Kenya. Parasites & vectors, 8, 496 vi). Microchip technology (presence of nucleic acid) vii). Use of salivary with an emphasis on rapid detection of infection by using point-of-care devices. Oral mucosal transudate contains secretory immunoglobulin (Ig) A, as well as IgM and IgG. Else by < Corstjens, P., L., William, R., A., and Malamud, D., Detecting Viruses by Using Salivary Diagnosis, J Am Dent Assoc. 2012 Oct; 143 (10 Suppl): 12S-18S > --Methods for Serodiagnosis (done in labs): i). Neutralization ii). Hemagglutination inhibition (HAI) iii). Complement fixation iv). Enzyme immunoassay (IgG or IgM) v). Western blot vi). Latex agglutination vii). Immunochromatography --Detection rates in comparison versus economic costs (money from use of tools and personnel, time). --Identify growth rate of respective virus for common parameter value ranges; may have to observe different geometric exhibitions concerning the pending combinations of variation ranges with the parameters. To then from sources to determine which methods provide best detection if group of viruses considered was extended (land ecosystems, aquatic, sustenance). Will incorporate competent use of microbial, viral, pathogen and EPA software from software portfolio. Can be compared with curve fitting of data, and regression modelling can also be done. --The same can be done with bacteriology. 10. Advanced identification of bacteria, fungi and viruses (laboratory activities). Activity 9 is a prerequisite. --Identification of bacteria (including mycobacteria) is based on growth characteristics (such as the time required for growth to appear or the atmosphere in which growth occurs), colony and microscopic morphology, and biochemical, physiologic, and, in some instances, antigenic or nucleotide sequence characteristics. The selection and number of tests for bacterial identification depend upon the category of bacteria present (aerobic versus anaerobic, Gram-positive versus Gram-negative, cocci versus bacilli) and the expertise of the microbiologist examining the culture. Gram-positive cocci that grow in air with or without added CO2 may be identified by a relatively small number of tests. The identification of most Gram-negative bacilli is far more complex and often requires panels of 20 tests for determining biochemical and physiologic characteristics. --The identification of filamentous fungi is based almost entirely on growth characteristics and colony and microscopic morphology. --Identification of viruses is usually based on characteristic cytopathic effects in different cell cultures or on the detection of virus- or species-specific antigens or nucleotide sequences. Will incorporate competent use of microbial, viral, pathogen and EPA software from software portfolio. Can compared with curve fitting of data, and regression modelling can also be done. 11. Biofilms experimental research Environmental signals and regulatory pathways that influence biofilm formation. Concerns as well survival, expansion, reduction and transferral in various environments and circumstances such as (i) Soil (ii) Aquatic (iii) Extreme environments (iv) Hosts (v) Ecological significance of plant-associated (vi) Bioremediation (vii) Waste water treatment systems (viiii) Corrosion & fouling (ix) Importance for the maintenance and monitoring of freshwater health (x) Extracellular Enzymes in Aquatic Biofilms: Microbial Interactions Vs Water Quality Romaní, A. M., Artigas, J., and Irene Ylla, I., Extracellular Enzymes in Aquatic Biofilms: Microbial Interactions vs Water Quality Effects in the Use of Organic Matter, Microbial Biofilms: Current Research and Applications (Edited by: Gavin Lear and Gillian D. Lewis). Caister Academic Press, U.K. (2012) Will incorporate competent use of microbial, viral, pathogen and EPA/USDA software from software portfolio. Can be compared with curve fitting of data, and regression modelling can also be done. NOTE: dental plaque and shower curtains may be excellent sources of biofilms. 12. Experimental investigation of the following article: Liu, W., et al, Ascorbic Acid Induces Cardiac Differentiation of White Adipose Tissue-derived Stem Cells, Molecular and Cellular Biochemistry (2019) 450: 65 – 73 However, preferably no surgical means on animals, instead to find other means of acquiring such cells. Interest in expanding investigation with various animal and humans. 13. Experimental investigation of the following article: Prasad, K., Is there any evidence that AGE/sRAGE is a universal biomarker/risk marker for diseases? Molecular and Cellular Biochemistry (2019) 451: 139 – 144 Identify other “universal” biomarkers/risk markers for diseases. Identify the models, formulas or quantities for determinations. Experimentally investigate as well and observe/determine whether alternatives are consistent with (standard) determination like what is expressed in above journal article. 14. Experimental investigation of the following article: Chanphai, P., Tajmir‑Riahi, H. A., Encapsulation of Micronutrients Resveratrol, Genistein, and Curcumin by Folic Acid-PAMAM Nanoparticles, Molecular and Cellular Biochemistry (2018) 449:157–166 Be economic as possible. Some of the mentioned substances are OTCs in pharmacies. Encapsulated versus naked may be interesting 15. Smartphone to Digital Microscope Conversion Materials: 3x 4 ½” x 5/16” carriage bolts 9x 5/16” nuts 3x 5/16” wing nuts 5x 5/16” washers ¾” x 7” x 7” plywood -- for the base ⅛” x 7” x 7” plexiglass -- for the camera stage ⅛” x 3” x 7” plexiglass -- for the specimen stage Scrap plexi (~ 2"x 4") for specimen slide (optional but useful) laser pointer focus lens (use at least two for increased magnification) LED click light (necessary only for viewing backlit specimens) Tools: Drill Assorted bits Ruler Note: one must acquire the lenses from laser pointers; cheap ones will do. Unlike how the lenses are fixed in place as shown in link, towards increasing certainty that smartphone isn’t compromised physical add-ons, one can possibly create a surface chamber for the two lenses in a means where the smartphone camera can be synchronized well on top of the lenses. If there is a better alternative to wood for the base, then so be it. Note: nobody cares what brand of smartphone you have as long as it has decent view. https://www.instructables.com/id/10-Smartphone-to-digital-microscope-conversion/ Such an apparatus must be sanitize capable where lenses and plexi glass will not be damaged; for metals in foundation corrosion resilience is will appreciated. Such a contraption with smartphone is easily portable for field observation and to serve as a financial backup when traditional microscopes are unavailable. There will be live recordings for various durations for various interests and to be archived. Play times must have the ability to be highly accelerated to one’s choosing without compromising the recordings. Note: this will be one of the first activities a student will do. One can readily take microscopic view photos and projector video recordings for data and archives. 16. H2O2 Cellular Signalling H2O2 is well known as a disinfectant both towards the skin and oral applications, and bleaching. Now, to investigate the role of H202 at the intracellular level. --Literature journal articles: Gough, D. R., and Cotter, T. G., Hydrogen Peroxide: A Jekyll and Hyde Signalling Molecule, Cell Death & Disease volume 2, page 213 (2011) Sies, H., Hydrogen Peroxide as a Central Redox Signalling Molecule in Physiological Oxidative Stress: Oxidative Eustress, Redox Biology 2017 Apr; 11: 613–619 Lennicke, C., Rahn, J., Lichtenfels, R., Wessjohann, L. A., & Seliger, B. (2015), Hydrogen Peroxide - Production, Fate and Role in Redox Signalling of Tumour Cells, Cell Communication and Signalling: CCS, 13, 39. --Biochemical history, Industry Standing and experimentation parameters: National Center for Biotechnology Information (2020). PubChem Compound Summary for CID 784, Hydrogen peroxide. --Journal Articles for developing experiments: Huang, B. K, and Sikes, H. D., Quantifying Intracellular Hydrogen Peroxide Perturbations in Terms of Concentration, Redox Biology 2 (2014) 955–962 Tomalin, L. E. et al, Increasing Extracellular H2O2 Produces a Bi-Phasic Response in Intracellular H2O2, with Peroxiredoxin Hyperoxidation Only Triggered Once the Cellular H2O2-Buffering Capacity is Overwhelmed, Free Radical Biology and Medicine 95 (2016) 333–348 Experimentation journal articles concern investigative experimentation towards comparative findings. 17. Comprehending how microbial enzymes function to drive ecosystem nutrient availability Exploring how key enzymes made by soil microbes function in the soil environment. They are important, because they induce the decay of organic compounds in soil that contain nutrients important for microbial and vegetation functioning. As such, they ultimately provide a large fraction of any ecosystem’s nutrient demand. We do not know how many of them respond to temperature, soil pH, soil mineralogy, or moisture availability. A set of projects exploring these questions. Will incorporate competent use of microbial, viral, pathogen and EPA software from software portfolio. Can be compared with curve fitting of data, and regression modelling can also be done. 18. Plant Biochemistry & Phytochemistry Properties of phytochemicals in plants, trees, fruits and vegetables...including all taxonomic distributions. Phytochemical knowledge and experimental labs about the natural source, classification, detection, extraction, isolation, nutritional, pharmacological and toxicological effects. Discriminate samples from different places for a respective plant by significantly distant residencies from each other. If plants used in journal articles are inaccessible, then substitute with local revered plants. There must be integrity with data findings (specimen identification, specimen samples, ambiance). Ambiance concerns Customary temperature ranges Sunlight exposure Water supply Soil constituents PH Microbial residents General ecology Identified chemicals, compounds, etc., will be cross-referenced with professional databases which will include uses, properties, etc. Findings can possibly be integrated into a GIS. Data will be securely archived and developed in a manner to discriminate from future data. However, data must be integrable with computational tools for different models, parameters estimations, trends, etc. Journal article example: (I). Lab development Senguttuvan, J., Paulsamy, S., & Karthika, K. (2014). Phytochemical analysis and evaluation of leaf and root parts of the medicinal herb, Hypochaeris radicata L. for in vitro antioxidant activities. Asian Pacific journal of tropical biomedicine, 4(Suppl 1), S359-67. Multiple plants/trees will be observed (including sorrel buds) besides what is observed in sources and articles (whether accessible or not). A supporting text for intelligence and experimentation practices Harborne JB 1998. Phytochemical methods. Chapman and Hall Pharmacognosy, Phytochemistry, Medicinal plants By Jean Bruneton (1995), English edition. Levoisier Publishing, Pari Mandatory range of interest: Sugars, soluble fibers and organic acids Fats and oils, Carotenoids Flavonoids, anthocyanins and polyphenolics Antiseptics Steroids Alkaloids and Seed storage proteins May extend to natural pesticides and cleaning agents For such range of interest students will be pursue determination of the metabolic process for each. Will also make use of biochemical software for amination models with properties and simulation of conventional chemical reactions. (II). The increase in antibiotic resistance bacteria and synthetic drugs has urged the search of new antibacterial and antioxidant agents from medicinal plants. To study the antibacterial activity and antioxidant activity of leaf extract of Andrographis paniculata. In this study, the powdered leaves are subjected to sequential Soxhlet extraction by increasing polarity index of solvents (hexane, chloroform, ethyl acetate and methanol). Prove or disprove: The methanol extract gave the highest percentage yield of extraction in the sequential extraction. All the solvent extracts were then used in phytochemicals screening tests, antioxidant and antibacterial assays as well as thin layer chromatography analysis. Prove or disprove: for the phytochemicals screening test, terpenoids was found to be the most abundant compounds in chloroform, ethyl acetate and methanol extracts. The DPPH assay was carried out to determine the antioxidant activity of all solvent extracts. Prove or disprove: Hexane extract found to exhibit the highest antioxidant activity with the lowest half maximal inhibitory concentration, IC50 value of 2.80 mg/ml. The antibacterial activity was evaluated qualitatively through agar disc diffusion toward Staphylococcus aureus, Staphylococcus epidermidis and Escherichia coli. Prove or disprove: the ethyl acetate extract showed the highest zone of inhibition value (17.0 mm) in S. epidermidis treatment. Prove or disprove: the potential of antibacterial activity decreases with chloroform and hexane. The methanol extract was failed to exhibit any antibacterial activity. Prove or disprove: all the solvent extracts showed no antibacterial activity against E. coli. The antibacterial activity is evaluated quantitatively through minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) tests. Prove or disprove: the MIC values of the ethyl acetate and chloroform extracts for S. aureus and S. epidermidis were ranged from 375 µg/ml to 750 µg/ml. Both solvent extracts were bacteriostatic toward the S. aureus and S. epidermidis. Prove or disprove: In conclusion, high extraction yield does not indicate that it has high antibacterial and antioxidant activities. Prove or disprove: The ethyl acetate extract performs the best antibacterial activity and hexane extract perform the best antioxidant activity. One may find simple elements as well. Concerning elements, molecules, compounds, etc. found in plants/trees to identify the role of each (if any) for metabolism, regulation, biochemical mechanisms, etc. 19. Ecological and evolutionary feedbacks between plants and beneficial fungi Lab investigation of the ecological and evolutionary consequences of plant-microbe interactions. Much of our work focuses soil fungi (called mycorrhizal fungi) that commonly improve plant growth through increased access to soil resources. We are particularly interested in how the benefits that plants receive from these fungi change over time and the potential role of these fungi in maintenance of plant species diversity. We often work in native prairie, where we have found that these fungi play primary roles in plant dynamics and in the restoration of native diversity. Work on this problem could involve work in the field, greenhouse, lab (at microscope work or DNA analysis) or computer (modeling). Will incorporate competent use of microbial, viral, pathogen and EPA software from software portfolio. Can be compared with curve fitting of data, and regression modelling can also be done. 20. Evaluation of natural repellents Sumangala Bhat, K. and Aravind G., Evolution, Current Status and Prospects of Phyto-Repellents against Mosquitoes, International Journal of Pharmacology, Phytochemistry and Ethnomedicine, Vol. 8, pp 54-73, 2017 https://www.scipress.com/IJPPE.8.54.pdf To analysis the given article and replicate experimentation to the best as possible. Specimen mosquitoes with controls and concealment required. 21. Parasitic Analysis Phase 1 (Urine testing)-- Activity concerns parasite findings or determination by urinary tests. If applied chemicals, substances in journal articles and sources are in accessible, then substitute with local economic alternatives. Will also incorporate professional microbiology and medical databases to make conclusions about findings. Multiple samples are required. Sources and journal article examples to emulate for experimentation: https://www.cdc.gov/dpdx/diagnosticprocedures/other/urine.html Lodh, N. et al, Diagnosis of Strongyloides stercoralis: Detection of Parasite-Derived DNA in Urine, Acta Tropica 163 (2016) 9–13 Will not be restricted to identification or sole pursuit of organisms encountered in sources and journal articles. Research host-parasite relationships in urinary tract infections. Based on observations or findings from urinary tests are there any coinciding findings with your research on host-parasite relationships in urinary tract infections? Phase 2 (Stool Examinations)-- The stool will be checked for colour, consistency, amount, shape, odour, and the presence of mucus. The stool may be examined for hidden (occult) blood, fat, meat fibers, bile, white blood cells and sugars called reducing substances. As well, the pH of the stool also may be measured. A stool culture is done to find out if bacteria may be causing an infection. Journal article example: Koontz, F., and Weinstock, J. V., The Approach to Stool Examination for Parasites, Gastroenterology Clinics of North America, Volume 25, Issue 3, 1 September 1996, pages 435-449 Will also incorporate professional microbiology and medical databases to make conclusions about findings. Note: for the same hosts urinary testing and stool testing can be cross-compared to observe which is more sensitive, or accurate. Phase 3 (other types of testing)-- Will also identify other types of tests and determine their economic standing. May proceed with experimentation (time permitting and if economically feasible). Will also incorporate professional microbiology and medical databases to make conclusions about findings, if done. 22. Infiltration and neutralization of microorganisms by biochemcal means (i) Peptidoglycan Biosynthesis Animation Make use of journal articles as sources as well. Phases warrant lab activities as well. Assist: --> Peptidoglycan Biosynthesis [HD Animation] - YouTube (ii) Apart from E coli will identify the unique compounds and/or processes involved with other chosen types of bacteria. (iii) Will then analyses and simulate the (bio)chemistry of antibiotics towards Peptidoglycan and bacterial DNA replication. Concerning molecular exhibitions and simulations various software have been mentioned throughout. Will try to identify why some antibiotics are more effective than others concerning Peptidoglycan and bacterial DNA replication; accompanying analysis molecular exhibitions and simulations for such reasons should also be developed. (iv) If one can model rate of effectiveness for each chosen antibiotic such would be nice. Of consequence environmental conditions can have influence on performance of antibiotic and welfare of different bacterial, respectively. Conditions examples are temperature, Ph, salinity, ionic level, presence of other molecules, light interaction (different from thermal), dosage, etc. (v) Will compare the (bio)chemistry of antiseptics versus antibiotics towards Peptidoglycan, bacterial DNA replication and generate sterilization. Will try to identify how effective antiseptics still are concerning bacteria evolution/mutations; environmental conditions to accounted for as well. Activity can also be extended to treat virology. For viruses with envelops one must establish definitively the role of such envelops. Examples of functions: Protecting the RNA or DNA molecule(s) Evading recognition by the immune system Facilitating virus entry Note: non-enveloped viruses will also be treated. Entry, transmission, and exit pathways of the (101) viral families on the 2013 International Committee on Taxonomy of Viruses (ICTV) list. A article assist: Buchmann, J. P. and Holmes, E. C. (2015). Cell Walls and the Convergent Evolution of the Viral Envelope. Microbiology and Molecular Biology Reviews, Volume 79 Number 4 One may need to review the mechanism of cell penetration by viruses if needed. To establish the (bio)chemistry with antibiotics and antiseptics concerning such envelope functions (and treatment also for nonenveloped viruses). The activities phases detailed for bacteria will also be “emulated”. Activity can be further extended towards general microrganisms known to be harmful to humans and animalia of interest. 23. Protein & DNA Imaging NOTE: at this level we are not interested on experimentation with rats and such sorts of things, neither any pursuit of cancerous cells. Part I -Review of of the Watson-Crick Experiment -Strawberry DNA extraction is one of easiest means; same likely to be for other fruits and vegetables, however, such may not be applicable to the type of methodologies of other parts of this activity. Nevertheless. It’s a good kindergarten activity to star with towards DNA imaging (to be done). There may issues of preserving the extracted DNA for long periods. Will compare such “kindergarten” extraction means to extraction with Phenol-Chloroform and NaClO4. Will determine whether the latter two methods are practical for extraction of DNA from produce. If so, logistics, economics and detailed chemistry will be treated; likely, lab exercises for the latter two if methods are applicable. Will then move on the human DNA extraction will the latter two methods, where respective logistics, economics and detailed chemistry will be treated. Will apply direct Raman spectroscopy to determine make up and, recognition o functional groups and other things. Hopefully, window of stability of nucleotides is enough to directly apply the Raman spectroscopy which doesn’t destroy the sample or specimen, consider preparation that can be employed in order to apply such Raman spectroscopy scheme. Part II Raman Scattering Microscopy The following journal articles provide economic means of access to DNA imaging. --Zhang, X. et al, Label-Free Live-Cell Imaging of Nucleic Acids Using Stimulated Raman Scattering Microscopy, ChemPhysChem 2012, 13, 1054 – 1059 --Lu, F. et al (2015). Label-free DNA Imaging In Vivo with Stimulated Raman Scattering Microscopy, PNAS Volume 112 Number 37 --Wang, O. et al, Mechanisms of Epi-Detected Stimulated Raman Scattering Microscopy, IEEE Journal of Selected Topics IN Quantum electronics, Vol. 18, NO. 1, January/February 2012 Proceed with development of active imaging. Part III Use of bright-field and ultraviolet (UV) induced fluorescence modes Abstract: “Imaging protein crystals and distinguishing them from salt crystals is an important task for protein crystallographers. The conventional tool used for this purpose is a dual-mode microscope composed of bright-field and ultraviolet (UV) induced fluorescence modes. The distinction between a protein and a salt crystal is made based upon the fluorescence response to the UV excitation, where most protein crystals absorb the UV excitation and emit fluorescence, unlike salt crystals. These dual-mode optical microscopes are sensitive; however, they are relatively bulky and expensive as they require UV-grade optics. As an alternative, here we demonstrate that on-chip UV holographic imaging offers a low-cost, portable, and robust technique to image and distinguish protein crystals from salt crystals, without the need for any expensive and bulky optical components. Only composed of a UV light-emitting-diode at 280 nm and a consumer-grade complementary metal–oxide–semiconductor image sensor decapped and interfaced to a Raspberry Pi single-board computer, the necessary information from the crystal samples (placed very close to the sensor active area) is captured in the form of in-line holograms and extracted through digital back-propagation. In these holographic amplitude reconstructions, protein crystals appear significantly darker compared to the background due to the strong UV absorption, unlike salt crystals which do not show any contrast, enabling us to clearly distinguish between them. We believe that the on-chip UV holographic microscope could serve as a low-cost, sensitive, and robust alternative to conventional lens-based UV-microscopes used in protein crystallography.” Journal article --> Daloglu, M. U. et al (2019). Low-Cost and Portable UV Holographic Microscope for High-contrast Protein Crystal Imaging. APL Photon. 4, 030804 Will develop all such in a lab for active imaging Part IV Immunofluorescence Methods What are such methods? Identification of the various uses of immunofluorescence Methods A major accomplishment will be imaging DNA or RNA or proteins; that would be a tremendous start. Then Lab experimentation similar to what is analysis in the following guides to be pursued. Analysis of biochemistry and organic chemistry for functional and competent activities is mandatory; accompany with visualization and simulation software expressed in many cases before. Optics and physics however for biological sciences students may be too technical and elusive for them. Imaginig labs will be done. --Boutorine, A. S. et al. Fluorescent Probes for Nucleic Acid Visualization in Fixed and Live Cells. Molecules 2013, 18, 15357-15397 --Pollex T, Piolot T, Heard E. Live-cell imaging combined with immunofluorescence, RNA, or DNA FISH to study the nuclear dynamics and expression of the X-inactivation Center. Methods Mol Biol. 2013;1042:13-3 --Kishi, J.Y., Lapan, S.W., Beliveau, B.J. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods 16, 533–544 (2019) --Bennett, B. T., Bewersdorf, J., & Knight, K. L. (2009). Immunofluorescence imaging of DNA Damage Response Proteins: Optimizing Protocols for Super-resolution Microscopy. Methods (San Diego, Calif.), 48(1), 63–71 --Smolka, J. A. et al. Recognition of Cellular RNAs by the S9.6 Antibody Creates Pervasive Artefacts when Imaging RNA:DNA Hybrids. bioRxiv 2020.01.11.902981 --Lin, J. et al. Highly Multiplexed Immunofluorescence Imaging of Human Tissues and Tumors using t-CyCIF and Conventional Optical Microscopes. bioRxiv 151738 24. Role of Oxidation-Reduction The ability of an organism to carry out oxidation-reduction reactions depends on the oxidation-reduction state of the environment. i. Consider the following journal article: Pezeshki, S. R., & DeLaune, R. D. (2012). Soil Oxidation-Reduction in Wetlands and its Impact on Plant Functioning. Biology, 1(2), 196–221 The pursuit is development of various model experimentation to confirm or disprove such article. Technicality and intricacy may go well beyond analytical knowledge and assumed time frame for operations. ii. Concerning oxidation-reduction will carry out environmental and laboratory investigation concerning the following parameters: << dissolved oxygen, alkalinity, biological CO2 pump, acidity, eutrophication, metal nutrition, metal pollution, excessive carbon emissions >> Will look at various land and aquatic environments concerning, microorganisms and vegetation. Definitive measures must be identified with data gathering towards conveyance of proof. Likely to have various test samples, test specimens, and trials. Will not necessarily seek extremes towards terminal cases of vegetation, but such is acceptable for microorganisms. As well, pursuit of analytical mathematical models relating various parameter, where field and lab investigation will be compared with such. Correlation analysis and causality determination can be incorporated. 25. Honey as a cloning agent Apart from the common knowledge will establish the comprehensive biochemical process involving DNA/RNA. Make use of journal articles and professional sources to support activity. Software can complement. Plant types to experiment with must naturally have high regeneration or growth rates. What are the similarities and differences between sophisticated conventional biological methods (SCBMs) and honey (as the cloning agent)? How effective is honey compared to the SCBMs? There must be experimental controls and trials to acquire significant statistical data? Is honey only applicable to plants? If not identify or favourable environments and process to fit the organism environment in question. Determine what other basic household “commodities” can be substitutes for honey. Establish the thorough biochemistry for such with appropriate processes and environments(s). Pursue experimentation with controls and trials as done with honey. 26. Reinforcement of Foundational Biochemistry Experiments Activity will assume successful completion of biochemistry II. Activity serves to reinforce basic biochemistry lab skills but will be a highly accelerated environment with experiments. Likely will be one of the earlier activities done (but not necessarily a prerequisite for various other activities). Experiment 1: Introduction to Techniques Use of pipetman Spectroscopy and dilutions Analysis of experiment 1 results Experiment 2: Protein Purification Purification of LDH LDH Enzyme assays Protein assays Calculation hints: Purification table Experiment 3: Characterization of LDH SDS PAGE Western blotting Gel filtration chromatography Protein crystallography Experiment 4: Enzyme Kinetics Km determination Lactate Km determination (continued) Pyruvate Km determination Inhibition kinetics Inhibitor type determination Chemical modification of LDH Experiment 5: Cloning of LDH PCR and plasmid preparation Agarose gels and restriction digests Ligation and transformation Selection and screening Screening and sequencing Activity measurements Other possible experiments: Amino Acid Composition of a Dipeptide pH Dependence of [whatever enzyme] Stereochemistry of the Fumarase Reaction — an NMR Study NOTE: proteins or other polymers of interest or choice can be substitutes in the future, or if more economic. 27. Inquiry-based Undergraduate Biochemistry Despite the following journal article being structured for a course, instead it will be in a setting like the other prior activities. Gray, C. et al (2015). Known Structure, Unknown Function: An Inquiry-based Undergraduate Biochemistry Laboratory Course. Biochemistry & Molecular Biology Education. Volume 43, Issue 4, pages 245 - 262 NOTE: various biochemical software from software portfolio are available to complement activity. 28. Knowledge and professionalism with dexterous biochemical software mentioned in software portfolio. Will identify major issues with observational and modelling of biochemicals, polymers and entities in molecular biology. Includes chemical and behavioural properties. Will have interactive investigation of how software (in the software portfolio) can resolve or assist with issues. Will also include inquiry on purpose or function of substances in question. Activity will not focus solely on proteins, yet decent guides for such: -Zacharias, Martin. Protein-Protein Complexes: Analysis, Modeling and Drug Design. Imperial College Press, 2010. Hence, activity may also creep into molecular modelling software --Dobson, Christopher M (2003). "Protein folding and misfolding". Nature. 426 (6968): 884–90 --Marsh JA, Hernández H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA (2013). Protein Complexes are Under Evolutionary Selection to Assemble via Ordered. Cell. 153 (2): 461–470 --Sudha, Govindarajan; Nussinov, Ruth; Srinivasan, Narayanaswamy. "An overview of recent advances in structural bioinformatics of protein-protein interactions and a guide to their principles". Progress in Biophysics and Molecular Biology. 116 (2–3): 141–50. --Marsh, Joseph; Teichmann, Sarah A (2014). Protein Flexibilty Facilitates Quaternary Structure Assembly and Evolution. PLOS Biology. 12 (5): e1001870 --Levy, Emmanuel D; Boeri Erba, Elisabetta; Robinson, Carol V; Teichmann, Sarah A (2008). Assembly Reflects Evolution of Protein Complexes. Nature. 453 (7199); 1262-5 NOTE: It’s not possible to be confined to a single software. This activity will be pushed early as possible, contingent upon students being recognised with background to manage activity. Activity will also be pushed to be repeated by students. Activity may or may not be done before activity 29. 29. Spectroscopy Methods Will be administrated by the Chemistry “department”. Outline for students in the biological sciences will be different to students in chemistry; chemistry students concern a foundation that’s more connected to physics, and the mathematics that involve such. The biological sciences out line will have no influence on chemistry students. See details under chemistry. 30. Foundation for clinical drug metabolism Part A The following two articles provided economical quality experimentation or drug metabolism. However, experiments can possibly incorporate other types of drugs (if able), but methods and/or strategies must be valid for them --> Juvonen, R. O., Pennanen, S. and Pasanen, M. A Convenient Laboratory Experiment for Teaching the Fundamentals of Drug Metabolism. American Journal of Pharmaceutical Education Vol. 61, Fall 1997 Zhang, D. et al (2012). Preclinical experimental models of drug metabolism and disposition in drug discovery and development. Acta Pharmaceutica Sinica B; 2(6): 549–561 Part B The following journal can provide a strong overview for cellular and molecular mechanisms of drug dependence (may actually be limited): Gupta, S., & Kulhara, P. (2007). Cellular and molecular mechanisms of drug dependence: An overview and update. Indian journal of psychiatry, 49(2), 85–90. For drugs experimented with in part A, without immediate confirmation by established professional data, to investigate methods or tools that are conventional or modern to identify target receptors, one’s vulnerability to drug dependence related to receptors, and possible toxic conditions. For those methods that are economically feasible and practical students to implement them, and compare results with established professional data. 31. Synthesizing Antibiotics PART A Making penicillin, ampicillin and other “off-shoots” of penicillin. Advanced stoichiometry will be heavily enforced. Chemical kinetics and energetics involved in processes. Various mentioned software in the “Goody Bag” post can assist. Spectroscopy tests (likely Raman) with multiple samples and comparison with professional databases. Tests on microbial, viral organisms. Will acquire specimen from various sources and cultured to substantial population. Generally, one has an ambiance where antibiotics are known to be behave optimally. Will like chronological imaging for a chosen duration. Penicillin and the “off-shoots” of penicillin will be tested against various cultures, where at least three sample tests per culture. Students will then have considerable theoretical development of large production by process design via simulators. Students must be creative with means of replicating environments possibly the acceleration of processes; possible hazards and volatilities identified. Accommodating simulation will be analytical quantitative modelling of systems; models and values have decisive role concerning materials, energy usage, quantity of products (and possible by-products), pollution, etc.. PART B Synthesizing sulfonamides. From the following link and journal articles will make great efforts towards the production of sulfonamides in the most economic and safe manner. Various mentioned software in the “Goody Bag” post can assist. Advanced stoichiometry will be heavily enforced. Chemical kinetics and energetics involved in processes. For sulfonamides there are many variants depending on the process applied. Spectroscopy tests (likely Raman) with multiple samples and comparison with professional databases. One will like multiple variables, hence difference processes of synthesizing. Tests on microbial, viral organisms. Will acquire specimen from various sources and cultured to substantial population. Generally, one has an ambiance where antibiotics are known to be behave optimally. Will like chronological imaging for a chosen duration. Likely each type of sulfonamide will be tested against various cultures, where at least three sample tests per culture--> https://www.organic-chemistry.org/synthesis/N1S/sulfonamides.shtm Reza Massah, A., Sayadi, S., & Ebrahimi, S. (2012). A green, mild and efficient one-pot method for the synthesis of sulfonamides from thiols and disulfides in water. RSC Advances, 2(16), 6606-6616. Naredla, R. R., & Klumpp, D. A. (2013). Preparation of sulfonamides from N-silylamines. Tetrahedron letters, 54(45), 5945–5947 Students will then have considerable theoretical development of large production by process design via simulators. Students must be creative with means of replicating environments, and possibly the acceleration of processes; possible hazards and volatilities identified. Accommodating simulation will be analytical quantitative modelling of systems; models and values have decisive role concerning materials, energy usage, quantity of products (and possible by-products), pollution, etc. PART C Antibiotics resistance modelling The following articles can be applied to antibiotics of part A and part B with a second stage of experiments, or to make use of the acquired data from the total process of the antibiotics administered towards modelling. Apart from differential equations will apply both linear regression, multilinear regression and possibly exponential regression. Make the adjustments to accommodate the antibiotics mentioned in part A and part B --> Spalding, C., Keen, E., Smith, D. J., Krachler, A. M., & Jabbari, S. (2018). Mathematical modelling of the antibiotic-induced morphological transition of Pseudomonas aeruginosa. PLoS computational biology, 14(2), e1006012 Bruce R. Levin, Klas I. Udekwu. Population Dynamics of Antibiotic Treatment: a Mathematical Model and Hypotheses for Time-Kill and Continuous-Culture Experiments. Antimicrobial Agents and Chemotherapy Jul 2010, 54 (8) 3414 – 3426 Jeffrey J. Campion, Patrick J. McNamara, Martin E. Evans. Pharmacodynamic Modelling of Ciprofloxacin Resistance in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy Dec 2004, 49 (1) 209 – 219 Jeffrey J. Campion, Philip Chung, Patrick J. McNamara, William B. Titlow, Martin E. Evans. Pharmacodynamic Modelling of the Evolution of Levofloxacin Resistance in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy May 2005, 49 (6) 2189 – 2199 PART D It’s only responsible that one investigates the performance of antibiotics in environments not optimal for respective antibiotic. That part will be somewhat and extension of part C, namely, modelling for environmental factors that can influence microbial populations subjected to antibiotics administered. Factors, such as sustenance, competition among species, temperature and so forth. One type of control may or may not be a culture without competition; there will be other controls in the experimentation process. Multiple regression will be employed for such a case with treatment of causal inference and possibility of lurking variables. 32. Biochemical tests In other actives samples of blood, urine and saliva are gathered to determine cases of infections. In this activity. Biochemical tests will be conducted in a hospital or health clinic to find out the individual’s status health. These biochemical test results are used by medical practitioners to perform diagnosis, prognosis and to monitor the disease. Kidney function test (Renal Profile Test) -The kidneys have important roles in regulating the electrolytes balance and acid-base homeostasis in the human body. The kidneys also monitor the body’s blood pressure and involved in excretion of waste products through urine and reabsorbing some substance that needed by the body. Kidneys also are able to filter the metabolic products such as urea and creatinine. Parameters tested in kidney function tests are sodium, potassium, urea and creatinine. -Sodium is the major electrolyte in water regulation in the body. No special preparation is required, and the patient’s blood can be taken at any time to run the test. Normal serum sodium levels are between 135-145 mmol/L. Hypernatremia is defined by an elevated sodium level in the blood while hyponatremia is a condition that occurs when the level of sodium less than 135 mmol/L. -Potassium helps nerves and muscles communicate. It also helps move nutrients into cells and waste products out of cells. The normal potassium level in the blood is 3.3-5.3 mmol/L. Hyperkalaemia is an excessive level of potassium in the bloodstream while hypokalaemia is a condition that occurs when the level of potassium is abnormally low. -Urea and creatinine have been used by clinician as a marker of renal function. Urea is a waste product formed from the breakdown of proteins while creatinine is a specific product for muscle metabolism. Increase concentration of urea in the blood may indicate acute and chronic renal dysfunction meanwhile increase creatinine level may indicate chronic renal dysfunction. A normal range for urea is 2.5 – 8.0 mmol/L while for creatinine is 62-106 umol/L (men) and 44-80 umol/L (women). Concerning the means to measure levels of sodium, potassium, urea and creatinine respectively, students will thoroughly establish the biochemistry analysis involved for credibility of such tests. Are kidney tests also good indicators of abnormalities or diseases of the nervous system? Pursue research that supports or refutes such. Lipid Profile test -Triglycerides are a type of fat found in the blood that the body uses for energy. Mostly, triglycerides are made in the liver and some are come from the diet. High triglyceride levels are associated with coronary heart disease. Many factors affect blood triglyceride levels including less physical activity, smoking, excessive alcohol consumption, intake of high carbohydrate diet, medication and some types of diseases and genetic disorders. The normal range for triglycerides is between 0.5 – 1.8 -Cholesterol is a type of lipid that is found throughout the body. It is produced by the liver and can be get from certain foods. Cholesterol is required in the body’s metabolic processes for the formation of new cells and synthesis hormones. However, excess cholesterol will form plaque between layers of artery walls, making it harder for heart to circulate blood. Thus, a clot blocks an artery will causes a stroke and heart attack. Hypercholesterolemia is a condition that occurs when the cholesterol level is high. A normal range for cholesterol is between 3.9 – 5.5 mmol / l. -Low density lipoprotein (LDL) contains mostly 75% of cholesterol and a smaller proportion of protein. LDL is responsible for delivering cholesterol to the parts of your body that need it. Excess LDL, however, causes a build-up of cholesterol in the walls of your arteries, contributing to the development of atherosclerosis. It is also known as a bad cholesterol. The normal range for LDL is <3.3 mmol/L. -High-density lipoprotein (HDL) is known as a good cholesterol. The job of HDL is to remove excess cholesterol from the cells and the walls of the arteries and then transport the cholesterol back to the liver for disposal. It may actually slow or even reverse the development of atherosclerosis. The normal range for HDL is between 1.0 – 2.2 mmol/L. Concerning the means to measure levels of Triglycerides, Cholesterol, LDL and HDL respectively, students will thoroughly establish the biochemistry analysis involved for credibility of such tests. As well does test for cholesterol count distinguish between good and bad cholesterol? If not, how is such done? Glucose test Blood glucose test measures the amount of a sugar called glucose in a sample of blood and this test may also be used to screen a person for diabetes. There are several types of glucose test included: -Fasting Blood Sugar (FBS). FBS are measured by taking a blood test after a period of fasting, usually of 8 hours without food. The normal range is between 3.0 – 5.4 mmol/L. -Random Blood Sugar (RBS). A blood sample will be taken at a random time to get a picture of the glucose concentration in the bloodstream. High random blood sugar will show that a person may has diabetes. But it is not a confirmatory test to diagnose a person with diabetes or not. The normal range is between 3.0 – 7.7 mmol/L. -2- Hour Postprandial Glucose (2HPP). This test measures blood glucose exactly 2 hours after eating a meal timed from the start of the meal. This test is done to see the effectiveness of the pancreas produces insulin to control the sugar. -Oral Glucose Tolerance Test (OGTT). The test is used to determine whether the body has difficulty metabolizing intake of sugar/carbohydrate. It is a screening test for the diagnosis of diabetes among pregnant women. The patient is asked to take a glucose drink (75g glucose in 250-300ml of water) and their blood glucose level is measured before and 2 hours after the sugary drink is taken. Uncontrolled diabetes can lead to complications of eyes, kidneys and heart. Concerning each type of glucose test, students will thoroughly establish the biochemistry analysis involved for credibility of such tests. Based on all such glucose results can one acquire a valid gauge on an individual’s activeness patterns, and their rate of fat burn? Liver function tests -Liver function tests (LFT) measure the levels of certain enzymes and proteins in the blood. This test is used to help diagnose and monitor liver disease or damage. LFT also used to assess the general state of the liver and it can also indicate other diseases, such as malnutrition or bone disease. -Alanine transaminase (ALT). Large amounts of ALT are found in liver cells. ALT is an enzyme that helps to process proteins. When the liver is injured or inflamed (as in hepatitis), the blood level of ALT usually rises. The normal range of values for ALT is between 0-41U/L. -Aspartate transaminase (AST) AST is an enzyme that plays a role in alanine and amino acid metabolism. When body tissue or an organ such as the heart or liver is diseased or damaged, additional AST is released into the bloodstream. The amount of AST in the blood is directly related to the extent of the tissue damage. The normal range of values for AST is between 10-35U/L. -Alkaline Phosphatase (ALP). Alkaline phosphatase (ALP) is an enzyme found in all body tissues especially in the liver, bile ducts and bone. ALP levels are higher than normal levels indicate the occurrence of cholestasis. The normal range for ALP is between 35-104 U/L. -Albumin and Total Protein. Albumin and total protein are made mainly in the liver. It helps our body as carrier protein, fight against infection and other function. Lower than normal levels may indicate liver damage or disease. The normal range for albumin is between 35-52 g/L for adults, while the normal range for total Protein is 66-87 g/L. -Total Bilirubin. Bilirubin is a brownish yellow substance found in bile. It is produced when the liver breaks down old red blood cells. Bilirubin is then removed from the body through the stool (feces) and gives stool its normal colour. Higher than normal levels of bilirubin may indicate different types of liver problems. Occasionally, higher bilirubin levels may indicate an increased rate of destruction of red blood cells. Increased levels of bilirubin can also be viewed with the naked eye, namely skin and eyes turn yellow. The normal range is between 0.0 – 17.1 umol/L. Concerning each type of enzyme test, Albumin -total protein test, and Bilirubin tests, students will thoroughly establish the biochemistry analysis involved for credibility of such tests. 33. Biosynthesis of Vitamins in Plants and Animals Main categorization Fat-soluble vitamins (A, D, E and K) Water-soluble vitamins (B and C) Note: for a particular vitamin in a category there are variants to consider, and such will be pursued. An example article guide: Fritsche, S., Wang, X., & Jung, C. (2017). Recent Advances in our Understanding of Tocopherol Biosynthesis in Plants: An Overview of Key Genes, Functions, and Breeding of Vitamin E Improved Crops. Antioxidants (Basel, Switzerland), 6(4), 99. Galmés, S.; Serra, F.; Palou, A. Vitamin E Metabolic Effects and Genetic Variants: A Challenge for Precision Nutrition in Obesity and Associated Disturbances. Nutrients 2018, 10, 1919. Such above guides only concern tocopherols, and rather, one will like guides for other vitamins and the variants concerning biosynthesis in plants and animals. Note: some variants may require particular conditions or creation with energy requirements and temperature requirements, and abundance of materials and catalyses, hence will detail such if needed. Will also make use of software to study and analyse different variants towards predicting use unique behaviours of variants towards nutrition, metabolism, reaction with other compounds (organic and inorganic), pharmaceutical, pharmacology, etc; some software also possess spectroscopy prediction tools (to compare with professional databases). Will also pursue possible intelligence on synthetic synthesizing of vitamins and the variants. If synthetic means are possible will develop process design towards simulation of production (and may be discriminating with variants if possible). May also pursue analysis of vitamins side effects and the conditions that lead to them; biochemistry will be thorough for all such, and will determine whether increased effects are dependent on variant type. 34. Mineral Regulation Concerns the major minerals and trace elements. Purpose in body function, appropriate forms and how such forms are attainable, intake, biochemistry of digestion and distribution for the various minerals, metabolic pathways. Includes detailed analysis of particular biological systems and corresponding cellular regulation, towards metabolic products, pathways with associated biochemistry. Consequences of deficiencies; reasons for possible negative side effects with intake; overdose. Will costruct processing stations similar to digestive and absorption bodily function. Stoichemetry, chemical thermodynamics, inorganic chemistry and so forth. Will also identify what activities lead to high reduction in mineral concentration in the body; must be specific with each mineral or trace element. 35. Advance Reinforcement of labs from the Bacteriology and Virology courses Competency and success in this activity will depend on retention and constructive effort in the Bacteriology and Virology courses. Skills acquired in such courses do have real value. Labs may be augmented a bit more. 36. Bacterial and Viral Nutrition PART A Bacteria types of interest: Autotrophs (different types) Organic compounds consuming bacteria Decomposers Heterotrophs that consume inorganic chemotrophs Accurate identification of different gathered specimen cultures from various environments is crucial. The following guide may or may not encompass all prior mentioned bacteria types concerning: Gottschalk G. (1986) Nutrition of Bacteria. In: Bacterial Metabolism. Springer Series in Microbiology. Springer, New York, NY Note: some aspects of activity will involve lab observations and record keeping, Microscopes with cameras that can capture culture behaviour towards projectors will be nice. Such microscopes with cameras should be able to observe bacteria physiology, take picture in high speed frames, and video record as well. Other observations and modelling include dynamic population evolution, growth rates, etc. alongside use of mentioned pathogen/microorganisms modelling software. Particular research, observation, data collection and analysis for: Physiology of digestion or respective autotrophic process Biochemistry of digestion or biochemistry of respective autotrophic process Duration for metabolic respective process Waste products Are mutations in bacteria highly influential on the biochemistry of nutrition and metabolism? Under what conditions or threshold doe a respective bacteria type become hazardous to other organisms (micro and multicellular). For multicellular effects for plants, invertebrates and invertebrates may be unique depending on quantities, time, etc. Particularly for bacteria that acquire nutrition by photosynthesis will like various different specimen cultures exposed to different electromagnetic wavelengths for data; determine what other environmental parameters that can be controlled and investigated alongside nutrition administering. For autotrophs that rely on chemicals there will be different specimen cultures exposed to different chemicals; determine what other environmental parameters that can be controlled and investigated alongside nutrition administering. For bacteria that consume organic compounds there will be different specimen cultures exposed to different compound groups; determine what other environmental parameters that can be controlled and investigated alongside nutrition administering. For bacteria that decomposed compounds there will be analogy to prior. For heterotrophs that consume Inorganic chemotrophs there will be analogy to prior. PART B Viruses are made of three major components (and establish what the other minor components are) Nucleic acids, which include DNA and RNA and they provide a set of genetic instructions for future viral reproduction. Protein coatings that protect the nucleic acids. Lipid coatings that surround the protein coatings, but this is not present in all viruses. Viruses with lipid coatings are called enveloped viruses as opposed to naked viruses. Viruses do not carry enzymes needed to carry out the chemical reactions for life. Instead, they carry only one or two enzymes that decode their genetic instructions. Thus, viruses depend on host cells for viral production and nutrient. Concerning nutrient acquisition, emphasizing thorough biochemistry, how is such process accomplished in detail? What is the active physiology during such process? Terms that may be of interest Lytic cycle Lysogenic cycle Note: electron microscope may not be accessible in abundance, however this activity part will still be pursued. Some aspects of part A are applicable or can be parallel, while others are not; figure such out. PART C May also treat other non bacteria microorganisms, which would lead to a comprehensive study for nutrition in microbiology. 37. Botanical Hormones (I). Types (and likely each type will have variation) auxins gibberellins ethene Other examples: http://www.cannagardening.com/plant_hormones (II). Genes that are responsible for such and reasons for dominant and recessive traits. (III). Creation/metabolism of such hormones and pathways. Will pay close attention to what functional groups in such hormone compounds are responsible for influencing ideal functions of respective hormone in ideal biological environments. Will also incorporate use of software to assist with biochemical molecular modelling, where some of such software provides prediction for spectroscopy characterisation (to be compared to professional databases). (IV). How are hormone concentrations regulated along with the biochemistry for such? (V). How are hormones stored? Will also make use of software to assist with such biochemical molecular modelling to recognise the storage states and the associated bonds in sugars and fats, and what chemical reactions will lead to the synthesis from such storage. (VI). Pursuit of isolation and extraction of plant hormones. An example text that may be of use: Koshiba T. (2010). Plant Hormones. Methods and Protocols, 2nd edn. Annals of Botany, 105(4), viii. As well, pursue journal article guides for such that provide highly economic means. (VII). Will recognise commercial usage of plant hormones and inquire about how they are manufactured on large scales. Based on (vi) and assuming there are methods to preserve such hormones outside of ideal biological environments, will like comparative spectroscopy analysis between hormones extracted based on (vi) and commercially manufactured samples; will like means to definitely identify hormones without destroying samples or large quantities. (VIII). Experiments with hormones on plants based on (VII) may or may not be feasible due to time constraints with botanical development. If feasible one must consider what environmental controls are to be administered among various trials. Regression models can be applied to data and causal inference and lurking variables. (IX). The latter 2 of the 3 mentioned hormones are influential on the ripening and spoiling of fruits. Sterilized fruits with temperature and moisture experiments concerning metabolic and quantity levels of hormones can be feasible. (X). Clearly differentiate between regulation processes in single cellular/microorganisms and animalia hormone processes. What are the major gaps or significant differences? (XI). Can plant hormones be used for human medicinal purposes? Note: in the future one can pursue other types of hormones for functions such as immunity defence, competitor signalling, etc. 38. Protein Function Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Protein Function. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26911/ Will make use of chemistry, different biochemical software for simulations and computations, and lab experiments to confirm the 27- 29 significant statements from source above. A few of the major statements --> --All proteins bind to other molecules; significant features that make this universal. --The Details of a Protein’s Confirmation determine its chemistry --Sequence Comparisons between protein family members highlight crucial ligand biding sites --proteins bind to other protein through several types of interfaces --The binding sites of antibodies are especially versatile --Binding strength is measured by the equilibrium constant --Enzymes are powerful and highly specific catalysts --Substrate binding is the first step in enzyme catalysts --Enzymes speed reactions by selectively stabilizing transition states --Enzymes can use simultaneous acid and base catalysis --Lysozyme illustrates how an enzyme works --Tightly bound small molecules add extra functions to proteins From all encountered statements and topics will like to determine or develop a systematic method for protein identification and structure based on chemistry, software and experiments, unique to spectroscopy. May be given protein proteins to work on. Can functional groups be determined based on such developed methodology? After conclusion drawn by students, molecules will then be revealed and students will observe the spectroscopy of them from databases to determine to have a gauge on how accurate they are. Students in general will not be penalized or ridiculed as along as the applied chemistry, software usage and lab experimentation are competent and correct. How conclusive or effective is the developed methodology? How economically tasking is such methodology? Apart from analytical modelling, in labs or simulations among different molecules (and family of molecules) will like data fitting for --> Enzyme kinetics Binding energies (likely to vary with molecular bindings considered) Activation energy models (likely to vary with molecular bindings considered) Here are some of these major statements (there are crucial others in provided source) --> 39. Guide to the Expression of Uncertainty in Measurement (GUM) and transcendence Thoroughly identify and analyse GUM. Our goal is to develop a logistical framework that’s universal with any experimentation in science. Developing competence is quite important. Re-orchestrating some basic physics and chemistry labs students may encounter uncertainty treatment. Will like to extend to such particular labs with the analysis from part A. PART A Analysis from the following guides --> 1. Evaluation of measurement data — Guide to the expression of uncertainty in measurement — JCGM 100:2008 https://www.bipm.org/utils/common/documents/jcgm/JCGM_100_2008_E.pdf 2. Evaluation of measurement Data – Supplement to the “Guide to the Expression of Uncertainty in Measurement” – Propagation of Distributions using a Monte Carlo Method. JCGM.101: 2008 3. Barry N. Taylor and Chris E. Kuyatt (1994). Guidelines for Evaluating and Expressing the Uncertainty of NIST Measurement Results. NIST Technical Note 1297. 4. https://isotc.iso.org/livelink/livelink/Open/8389141 5. Ferrero, A., & Salicone, S. (2018). A Comparison Between the Probabilistic and Possibilistic Approaches: The Importance of a Correct Metrological Information. IEEE Transactions on Instrumentation and Measurement, 67(3), 607-620. Other applications ---Krouwer, J. (2003). Critique of the Guide to the expression of Uncertainty in Measurement Method of Estimating and Reporting Uncertainty in Diagnostic Assays. Clinical Chemistry, 49(11), 1818-21. ---Velychko, O., & Gordiyenko, T. (2009). The use of Guide to the Expression of Uncertainty in Measurement for Uncertainty Management in National Greenhouse Gas Inventories. International Journal of Greenhouse Gas Control, 3(4), 514-517. 40. Genome determination labs Software --> R and packages of interest (BiocManager, ChemmineR, dPCP, MAPITR) Thodberg, M., & Sandelin, A. (2019). A step-by-step guide to analysing CAGE data using R/Bioconductor. F1000Research, 8, 886 Cao, Y., Charisi, A., Cheng, L. C., Jiang, T., & Girke, T. (2008). ChemmineR: a compound mining framework for R. Bioinformatics (Oxford, England), 24(15), 1733–1734 < manuals.bioinformatics.ucr.edu/home/R_BioCondManual > < cran.r-project.org/web/packages/BiocManager/vignettes/BiocManager.html > Plant breeding and genomics < https://plant-breeding-genomics.extension.org/r-tutorials/ > PART A Brown T A. Genomes. 2nd edition. Oxford: Wiley-Liss; 2002. Chapter 7, Understanding a Genome Sequence. Available from: https://www.ncbi.nlm.nih.gov/books/NBK21136/ The mentioned outcomes from such above chapters of both texts are listed beneath, however, after reading further texts and articles may be required to analyse the intricate details and logistics. --Describe the strengths and weaknesses of the computational and experimental methods used to analyse genome sequences --Describe the basis of open reading frame (ORF) scanning, and explain why this approach is not always successful in locating genes in eukaryotic genomes --Outline the various experimental methods used to identify parts of a genome sequence that specify RNA molecules --Define the term ‘homology’ and explain why homology is important in computer-based studies of gene function --Evaluate the limitations of homology analysis, using the yeast genome project as an example --Describe the methods used to inactivate individual genes in yeast and mammals, and explain how inactivation can lead to identification of the function of a gene --Give outline descriptions of techniques that can be used to obtain more detailed information on the activity of a protein coded by an unknown gene --Describe how the transcriptome and proteome are studied --Explain how protein interaction maps are constructed and indicate the key features of the yeast map --Evaluate the potential and achievements of comparative genomics as a means of understanding a genome sequence Mentioned experiments that are deemed economic will be pursued in lab. Yeast will not be the only pursuit. There’s interest in botany, animal and human cells as well. There are various software, databases and clouds mentioned in the “Goody Bag” post to compare with. If needed the R environment with particular packages are verify useful. PART B Micro Array Analysis Micro array experiments will be pursued Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. Studying Gene Expression and Function. Available from: https://www.ncbi.nlm.nih.gov/books/NBK26818/ Taub, Floyd (1983). "Laboratory methods: Sequential comparative hybridizations analysed by computerized image processing can identify and quantitate regulated RNAs". DNA. Tarca, A. L., Romero, R., & Draghici, S. (2006). Analysis of Microarray Experiments of Gene Expression Profiling. American journal of obstetrics and gynecology, 195(2), 373–388 Jaksik, R., Iwanaszko, M., Rzeszowska-Wolny, J., & Kimmel, M. (2015). Microarray Experiments and Factors which Affect their Reliability. Biology direct, 10, 46 Bobashev G.V., Das S., Das A. (2002) Experimental Design for Gene Microarray Experiments and Differential Expression Analysis. In: Lin S.M., Johnson K.F. (eds) Methods of Microarray Data Analysis II. Springer, Boston, MA Júlio S. S. Bueno Filho, Steven G. Gilmour and Guilherme J. M. Rosa. Design of Microarray Experiments for Genetical Genomics Studies. Genetics, 2006 vol. 174 no. 2 945-957 Activity may go beyond genome detection. The following source provides various other applications that can be pursued --> https://en.wikipedia.org/wiki/DNA_microarray After readings, further texts and articles may be required to analyse the intricate details and logistics. There are various software, databases and clouds mentioned in the “Goody Bag” post to compare with. If needed the R environment with particular packages are verify useful. 41. Analysis, Logistics and experimentation for development of vaccines A. Basic questions --> Towards developing a vaccine, where does one start? What common methods or investigations are applied to do just that? Are vaccines more economic long term than antibiotics? Note: the adjective “economic” doesn’t only imply finance. B. Analysis of methods C. Logistics for chosen methods and investigations D. Will use very old and generic vaccines upon low threat microbes to confirm the vaccine research. One must clearly understand the applied investigation/process of prospective vaccine determination; will replicate/reproduce all such in lab. E. Testing vaccine Example sources to build activity upon --> --Detrick, B., Hamilton, R. G. and Schmitz, J. L. (2016). Manual for Molecular and Clinical Laboratory Immunology. ASM Books Series, ASM Press, 1240 pages --Grimes, S. E. A Basic Laboratory Manual for the Small-Scale Production and Testing of I-2 Newcastle Disease Vaccine. RAP publication 2002/22. FAO-APHCA --Rid, A. et al. (2014). Placebo use in vaccine trials: recommendations of a WHO expert panel. Vaccine, 32(37), 4708–4712 NOTE: will be using isolated tents. NOTE: for economic reasons chosen operations will be similar but not exact as described observations from example sources. NOTE: there may be ethical issues for some, so will try to reduce operations with elements of animal reproduction. NOTE: sterilization and annihilation of cultures or microbes after use of tools are essential; includes lab. Other things may need to be disposed of properly. 42. Guiding principles for monte carlo Analysis Students must successfully complete Biostatisitcs I & II to participate in this activity. Literature of concern --> Guiding Principles for Monte Carlo Analysis. Risk Assessment Forum U.S. Environmental Protection Agency, EPA/630/R-97/001: https://www.epa.gov/sites/production/files/2014-11/documents/montecar.pdf Literature provided is quite condense. A major goalis to develop a framework where all methods and analyses are sequential in a fluid, tangible and practical manner. Activity will be heavily reliant on data from labs and field activities. Activity will make extensive usage of R and RStudio. 43. Virology Labs Recital Activity will be based on the lab experience frm the following courses: Virology Tissue Culture & Virology Lab Hence, prerequisite will be students have taken at least the Virlogy course. Notes taken will be fast tracked and encountered labs will be replicated, however organisms may be different. Labs include use of all encountered software and journal articles. 44. Cyanobacteria Oxygen Production Involves multiple types of cyanobacteria in analysis and experimentation Environments: acquatic, quatic + ferrous A. Description of biochemical processes Will identify any uniqueness among the chosen types of cyanobacteria B. Modelling dcyanobacteria population/growth General analytical bacteria modelling Use of databases and software to refine prior For different types of cyanbacteria C. Investigtiong the role of the following variables on cyanobacteria growth and oxygen production Brightness UV Radiation Vacuum UV (100–200 nm) UVC (200–280 nm) UVB (280–315 nm) UVA (315–400 nm) Temperature Atmospheric Pressure Salinty < fresh to various levels of salinity > Acidity Carbon Dioxide concentration Nitorgen concentration What is hazardous to cyanobacteria gowth and stability? D. How do such prior variables in (C) influence our development in (B)? E. issue with toxins as by-products of cyanobacteria metabolism, and resolutions F. Cyanobacteria playing a crucial role in nitrogen fertilizer. Description of biochemical processes Will identify any uniqueness among the chosen types of cyanobacteria G. Coexistence with other organisms in environment Competition? (sustenance, terrain & competing biochemical processes) Mutualism? Detail biochemical processes H. Chloroplast being cyanobacterium living within plant cells. Research such. Following, how can we prove such with lab experiments? Lab experiments will be pursued. I. Developments from (A) through (H) will be applied to design and administer robust lab experiements. Additionally, the following literature can be very useful for activity: Salinity Intelligence --> Martin Hagemann, Molecular Biology of Cyanobacterial Salt Acclimation, FEMS Microbiology Reviews, Volume 35, Issue 1, January 2011, Pages 87–123 Shetty, P., Gitau, M. M. and Maróti, G. (219). Salinity Stress Responses and Adaptation Mechanisms in Eukaryotic Green Microalgae. Cells, 8, 1657 Pade, N., & Hagemann, M. (2014). Salt Acclimation of Cyanobacteria and their Application in Biotechnology. Life (Basel, Switzerland), 5(1), 25–49. General Intelligence --> Rantamäki S, Meriluoto J, Spoof L, Puputti EM, Tyystjärvi T, Tyystjärvi E. Oxygen produced by cyanobacteria in simulated Archaean conditions partly oxidizes ferrous iron but mostly escapes-conclusions about early evolution. Photosynth Res. 2016 Dec;130(1-3):103-111 Szeinbaum N, Toporek YJ, Reinhard CT, Glass JB. Microbial helpers allow cyanobacteria to thrive in ferruginous waters. Geobiology. 2021 Sep;19(5):510-520. Verseux, C., Heinicke, C., Ramalho, T. P., Determann, J., Duckhorn, M., Smagin, M., & Avila, M. (2021). A Low-Pressure, N2/CO2 Atmosphere Is Suitable for Cyanobacterium-Based Life-Support Systems on Mars. Frontiers in microbiology, 12, 611798. Kihara, S., Hartzler, D. A., & Savikhin, S. (2014). Oxygen Concentration Inside a Functioning Photosynthetic Cell. Biophysical Journal, 106(9), 1882–1889 45. Medical Drugs PART A: Drug Discovery Rather being a course, to operate as an activity. For mentioned tools and technologies it’s likely that generic versions can be substitutes. Fray, M. L. et al (2013). A Practical Drug Discovery Project at the Undergraduate Level. Drug Discovery Today Volume 18, Numbers 23/24 PART B: Drug Design for Undergraduates The given article is a basic guide towards drug design. Naturally, instructor can expand upon such article. Tantillo, D. J. et al (2019). Computer-Aided Drug Design for Undergraduates. Journal of Chemical Education, 96(5), pages 920 – 925 Further expansion: Sliwoski G, Kothiwale S, Meiler J, Lowe EW Jr. Computational methods in drug discovery. Pharmacol Rev. 2013 Dec 31;66(1):334-95 Concerning both prior articles any software specifically mentioned can possible be substituted by the following << Dalton, CP2k, Firefly, Gaussian, GAMESS-US, MOLDEN, NWchem, GPAW, Octopus, ORCA, FreeON, PUPIL, VOTCA, BOSS >> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream (http://bionano.cent.uw.edu.pl/software/) >> << TINKER >> Constituents of biochemistry/metabolic biology can also expand upon this activity with comprehensive pathway mapping and associated metabolic reactions throughout; possibly, also identifying side effects throughout metabolism. PART C: Drug Delivery Determine most constructive order: Farrell, S., Savelski, M., Hesketh, R. and Slater, C. S. (2006). Experiments in Drug Delivery for Undergraduate Engineering Students. American Society for Engineering Education Farrell, S and Hesketh, R. (2002). An Introduction to Drug Delivery for Chemical Engineers. Chemical Engineering Education http://users.rowan.edu/~hesketh/hesketh/cee%20drug%20delivery.pdf Farrell, S. and Vernengo, J. (2012). A Controlled Drug-Delivery Experiment Using Alginate Beads. Chemical Engineering Education, v46 n2 p97-109 Farrell, S. et al An Experiment to Introduce pH-responsive Hydrogels for Controlled Drug Delivery: Mechanical Testing. 120th ASEE Annual Conference and Exposition 2013
46. Chemistry of Flavour and Fagrances Engineering compounds or molecules to replicate taste types and smells. Obligations --> Memory Consolidation Include sensory and neural processes Some Key Areas in Development Considerable use of biochemistry and organic chemistry Modelling and Simulation software operations Hypotheses and means of validation Synthesizing processes and production models Stablization of compounds/molecules Behaviour with other substances and hazards analysis Means of safety testing Assisting texts --> Rowe, D. J. (2004). Chemistry and Technology of Flavours and Fragrance, WileyBlackwell Berger, R.G.. (2007). Flavours and Fragrances: Chemistry, Bioprocessing and Sustainability, Springer-Verlag Berlin Heidelberg Sell. C. S. (2006). The Chemistry of Fragrances: From Perfumer to Consumer. Royal Society of Chemistry NOTE: particular laboratory methods/techniques from texts will be pursued 47. Bioprocessing PART A Experiment 9 from the Food Chemistry course can apply here, but will be extended with temperature studies, and studies among various added yeast and added sugar types agents. Will also be pursuing enzymology operations. NOTE: activity will take on a serious bioprocessing tone, hence technical features such as models, modelling, parameters, quantities and measures will be emphasized. Other possible interests: mixed atmosphere packaging, pasteurization, and waste treatment. Software of interest R + R Studio Excel COCO (+ ChemSep), DWSIM (+ ChemSep) << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> PART B Insulin production with yeast Laboratory production of insulin by recombinant DNA technology with yeast population growth. Fermentation or whatever process to be coerced in a isolated environment; characteistics of environment must be particular and stable to acquire both optimal yeast growth and optimal insulin production. Satefy protocols and regulation with microbes Review of recombinant DNA technology Process and logistics for yeast with recombinant DNA technlogy Environmental conditions regulation For optimal yeast growth & optimal insulin production Identify & comprehend models, modelling, parameters & measures Biochemistry and Organic Chemistry modelling Analysis with possible by-products Stoichiometry Growth models based on whatever parameters prefrences Production expectation at time Tfinal Tools and structure for data gathering and assurance Description and profiling of the biprocessing system Means of insulin separation & gathering for storage & long term stablity Walk-through of operation procedures of bioprocessing system With logistical saftey protocols for whatever phases With tools and structure for data gathering and assurance Review of preparation for the relevant phases in development Implementation of bioprocessing Means of confirming insulin product Means of determining gross production and if consistent with preliminary modelling expectations Economics and innovation with technologies Software of interest R + R Studio Excel COCO (+ ChemSep), DWSIM (+ ChemSep) << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> A possible assist: Baeshen, N. A. et al. (2014). Cell Factories for Insulin Production. Microbial Cell Factories, 13, 141 48. Detection of Metabolites of Narcotic Drugs Activity may be a challenge to pursue due to legal grounds and policies of medical services. PART A Profiling or characteristion of illegal drugs (cocoaine, heroine, etc.) and opiods PART B Biochemical pathways for process aliments, disease. PART C Modelling and simulation of drugs (functional groups, active sites and reactions) subject to part B. Software use modelling and simulation will follow. The following are literature of interest throughout activity (but can be augmented by others): --Braithwaite, R. A. et al (1995). Screening for Drugs of Abuse. I: Opiates, Amphetamines and Cocaine. Annals of Clinical Biochemistry, 32(2), 123–153. --Simpson D, et al. (1997). Screening for Drugs of Abuse (II): Cannabinoids, Lysergic acid Diethylamide, Buprenorphine, Methadone, Barbiturates, Benzodiazepines and Other Drugs. Ann Clin Biochem. 1997 Sep;34 ( Pt 5):460-510. --Recommended methods for the Identification and Analysis of Fentanyl and its Analogues in Biological Specimens. UNODC -Busardo, F. P. et al (2019). Ultra-High-Performance Liquid Chromatography-Tandem Mass Spectrometry Assay for Quantifying Fentanyl and 22 Analogues and Metabolites in Whole Blood, Urine, and Hair. Front Chem. 7: 184 --Hazarika, P. et al (2010). Multiplexed Detection of Metabolites of Narcotic Drugs from a Single Latent Fingermark. Anal. Chem. 82(22), 9150–9154 --Pantano, F. et al (2017). Determination of Oxycodone and its Major Metabolites Noroxycodone and Oxymorphone by Ultra-High-Performance Liquid Chromatography Tandem Mass Spectrometry in Plasma and Urine: Application to Real Cases. Clinical Chemistry and Laboratory Medicine (CCLM), 55(9), 1324-1331. 49. Bacterial Toxins Purposes and stimuli Genome, genes, pathways, synthesizing, etc. Hopefully, one can create a contained “biosphere” having both bacterial and viral cultures (and possibly other types of microorganisms present), to observe the role of “toxins”. Analysis of conditions for stability. Neutralization. Dangers of “toxins” to humans Effects (ailments and diseases) Pathways Metabolic Pathways Conditions for toxin stability Transmission possibilities Neutralization Note: may also consider analogy for plant life welfare Literature assist --> Sharma, A. K., et al. (2017). Bacterial Virulence Factors: Secreted for Survival. Indian journal of Microbiology, 57(1), 1–10 Schmitt, C. K, Meysick K.C. & O'Brien A. D. (1999). Bacterial Toxins: Friends or Foes? Emerg Infect Dis. 5(2):224-34. Rudkin J. K., McLoughlin R.M., Preston A., Massey R. C. (2017) Bacterial Toxins: Offensive, Defensive, or Something Else Altogether? PLoS Pathog 13(9): e1006452. Harms, A. et al (2018). Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Molecular Cell 70, pp 768 – 784 Software for possible intelligence and profiling throughout development --> << COPASI, Pathvisio + Cytoscape + KEGG >> << Combase (Predictor and Modelling Toolbox) with MRV, USDA Pathogen Modeling Program, EPA Virulo >> << VCell, TiQuant + TiConstruct + TISIM >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> Software to be applied for modelling and simulation (mandatory): << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream (http://bionano.cent.uw.edu.pl/software/) >> << TINKER >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> For modelling and simulation, concerned with environments of high toxin functionality. The biochemical or molecular biological processes that lead to products, by-products and effects in the overall process (against other microorganisms and humans) with high functionality. Investigating neutralization resolutions as well. 50. Viroids and Prions (COMING SOON) Note: various lab sequences or sets from microbiology courses or metabolic biology can be extended or advanced to serve as activities; for the courses built upon they will be prerequisites however. Will have chosen lab sequences or sets from courses. Note: there’s the possibility of other projects. Other activities in Microbiology and Metabolism/Biochemistry are possible. There are other activities for Life Sciences students under Chemistry. Check such section. JOINT RESEARCH VENTURE POSSIBILITY Between the Oceanography and Biological Sciences (Oceanography, Microbiology, Metabolic Biology). Of interest are “Analytical Methods” and Research Publications” from the links: Method 544: Determination of Microcystins and Nodularin in Drinking Water Method 545: Determination of Cylindrospermopsin and Anatoxin-a in Drinking Water Method 546: Determination of Total Microcystins and Nodularins in Drinking and Ambient Waters Method for Determination of Cylindrospermopsin and Anatoxin-a in Ambient Freshwaters Method for Determination of Microcystins and Nodularin in Ambient Freshwaters Models of ecophysiology (PnET) Forest growth and carbon exchange (EDII) Forest landscape processes (Landis-II) Earth systems (CLM5) C. BOTANY NOTE: this degree pursuit will have one additional course more than the other degrees, but the ends will justify such investment. --Core Courses Scientific Writing I & II, General Biology I & II, General Chemistry I & II --Mandatory Courses Calculus for the Biological Sciences I & II, ODE, General Physics I --Mandatory Bridging Foundation Organic Chemistry I (with labs), Biochemistry (with labs), Organic Synthesis Laboratory, Cell Biology (with labs), Biostatistics I & II, Advanced Statistical Modelling and Machine Learning for Biostatistics, Geographical Information Systems --Mandatory Botany Foundation General Botany; Plant Symbioses; Field Botany; Plant Systematics; Plant Physiology; Dendrology; Aquatic Botany; Plant Nutrition; Plant Biochemistry; Functional Genomics; Plant Molecular Biology (with labs); Forest Ecology; Ecology Methods; Plant Pathology; Forest Pathology; Invasive Ecology; Plant Propagation I & II; Wildlife Conservation Models --Public Administration Internships (concerns no habitat destruction in forestry and conservation grounds) Botanical Gardens & Horticulture Forestry Ranger immersion Ecological Restoration Invasive Species Analytical Chemistry methods in various environments Substances Synthetics Pollution Contamination Pesticides Plantae Studies Biochemistry Nutrition Molecular Biology Genomics Pathogens and Diseases Biostatistics I Course concerns probability and statistics applied to problems in biology, industrial/occupational health, and epidemiology. Use of statistical software R for data analysis is emphasized extensively. Note: this course is designed for students majoring in the biological sciences with a second term calculus background. Through the extensive use of practical examples, this course is expected to motivate and teach students statistics knowledge that would be helpful for their major study. The computer program R is the standard statistical program for this course. Students will use R to complete data analysis projects. R can be downloaded and installed on your personal computer for free following instructions at http://www.r-project.org/. In addition, the R environment will be augmented by RStudio interface with other R packages. This course covers fundamental concepts in probability and statistics, including data description, design of experiment, probability rules and distributions, statistical inference and linear regression. Definitions will be learned through real-world examples and applications. Besides these traditional materials and subjects, topics and methods that are particularly applicable to the biological sciences will be introduced. Again, much focus on the applications of statistical ideas to realistic data and practices. Students are expected to use materials learned from this course to guide statistical practice for their major studies in the future. After successfully completing this course the expectation is that students will be able to: 1. To grasp concepts in probability and be able to apply basic probability rules, distributions, and laws to solve conceptual statistics questions 2. Use statistical guidelines and common sense to interpret the process of data collection, description and analysis, and to design statistically sound experiments 3. Learn various statistical inference techniques and be able to select appropriate methods for specific data sets and scientific purpose 4. Link the course materials with real-life examples, and explore the opportunities for other biological applications 5. Interpret statistical reports and carrying out data analysis using R. Several data projects will be assigned during the semester. Independent work is expected. This is not a course of “pen and paper finesse” succeeding the composition of gunk and bamboozle on a writing board. One can’t be successful in statistics by only writing down theory. Practice with an environment that applies intelligence and engagement is essential. There’s no point in doing statistics if one doesn’t know how to acquire and manipulate real data. Real data realistically outnumbers the fingers and toes one possesses. Most of grading will be based on projects (having commentary descriptions) accompanied by the analytical process development description done in a word processor. Typical Texts --> Will make use of R language Statistics texts under CRC Press and Springer publishing Tools --> R language and R Studio Note: a calculator at times may prove useful for the idealistic or the “synthetic” customary questions. “R Monograph Notebook” --> Students should maintain a notebook as they proceed through the course and learn how to do analyses in R. This assignment involves a notebook that lists the syntax and provides a brief explanation of each function that students learn during the course. Instructor will assign R maturity development questions to be in tune with course progress; you will only be allowed to use your monograph to assist with assigned questions. The notebook will be handed in near the end of the semester and handed back to the students after grading. Such a notebook can be an extremely useful resource both during and after the course to quickly refresh one’s memory on the details of a particular function. Design with R most likely will vary among students. Poor development in a such a notebook may or may not correlate with poor grades. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS. Grading --> Problem Sets 20% R Labs 25% R Activities 0.7 Will involve all course topics R Monograph Lab Notebook 0.3 R maturity development questions End of term lecturer observation 3 Exams 30% Assigned Projects 25% Exams --> Limited open notes. I don’t like setting up myself and students for embarrassment; you are not perfect with statistics, so expect exams to be primarily knowledge based and the calculus related fodder. Most of your development will come from homework and labs; it is what it is. Note: limited open notes. Students may be more comfortable with certain R packages. Again --> Several data projects will be assigned during the semester. Independent work is expected. Course Outline --> Introduction to statistics, data and R Statistical Measures and Summary Statistics for data sets Data acquisition: Sources/databases (ecological & biological), file types, APIs, etc. Data Wrangling Summary Statistics Applied probability theory Axioms of probability Modelling frequencies and establishing densities Simulating random variables from real experiments Probability distributions and properties Law of Large Numbers and the Central Limit Theorem Introduce the Law of Large Numbers (LLN) Central Limit Theorem (CLT). Identify Exponential, Poisson and Binomial data and respectively determine in a manner to confirm LLN and CLT. Routledge, R., Chebyshev’s Inequality, Encyclopaedia Britannica Is there too much reliance on assuming normal or Gaussian distribution? Towards Chebyshev’s inequality what amount of repetition (regarding LLN and CLT) of an experiment is adequate towards Chebyshev becoming relevant? Overview and goals of various concentration inequalities (just a survey). Sample Estimates Chi-square distribution The bottom line is to establish the flow of the uses competently with applications involving real raw data. Comprehending categorical data sets Organisation of data and sensitivity of categories concerning traits of interest. Test for independence McHugh ML. (2013). The chi-square Test of Independence. Biochem Med (Zagreb). 23(2): 143-9. Test of homogeneity Test of variance Applications of the Chi-Square distribution with confidence intervals T-distribution Kim T.K. & Park J. H. (2019). More About the Basic Assumptions of T-test: Normality and Sample Size. Korean J Anesthesiol. 72(4): 331-335. Sample size determination Population parameter estimation Confidence intervals Directly logistical to understand what you’re doing in R F-distribution Assumptions for the F-distribution Relevance to the biological sciences (active data immersion) Note: not textbook finesse, rather how and when actively. Goodness of fit: fit of distributions Summary Statistics Skew and Kurtosis P-P and Q-Q Statistical Tests Definition, Null hypothesis One-sided & two-sided tests of hypothesis Types of test statistics Comprehending critical values for ideal distributions Significance levels Critical values for real raw data sets Does your data exonerate ideal distributions? Chi-Square Test Kolmogorov-Smirnov Test Anderson-Darling test Shapiro-Wilk Test MLE and Method of Moments Manual tasks will be limited to at most 4 element data sets Computational logistics for large data sets followed by implementation Review/probe data for goodness of fit module for appropriate distribution You may be tasked with distribution determination before parameter/point estimation Hypothesis testing (exploratory, sans assumption of distribution) Note: as aspiring biologists I can’t give “zombie textbook problems” and expect you to relate to a profession tangibly and fluidly. You will be exposed to raw professional data from various sources. You will develop the four mentioned steps. You should ask yourselves if the hypotheses are practical as well. Why is normal distribution assumed? Will be exploratory rather than zombie problems. Namely, knowledge and skills from Goodness of fit module. Then proceed with the following: 1.State the two hypotheses so that only one can be right 2.Formulate an analysis plan, outlining how data will be evaluated 3.Carry out the plan and physically analyze the sample data 4.Analyse the results and either reject the null, or state that the null is plausible, given the data 5.Directly logistical to understand what you’re doing in R Test of Proportions (exploratory) Differences in population (mean and median) Comparisons of variances Confidence limits for means. Does it require normality? Correlation (includes misuse of types and resolutions) Pearson Correlation Crucial Conditions Structure Implementation Spearman Correlation Crucial Conditions Structure Implementation Kendall Correlation Generating heat maps. The ggpairs() function Bivariate Regression Multiple Regression Model components Methods to select variables OLS MUST: Summary Statistics and forecasting Analysis of Covariance Must be exploratory, else it’s toxic Non-parametric statistics Resampling methods Falsified Data Hartgerink C, Wicherts J, van Assen M (2016). The Value of Statistical Tools to Detect Data Fabrication. Research Ideas and Outcomes 2: e8860. Al-Marzouki, S., Evans, S., Marshall, T., & Roberts, I. (2005). Are these data real? Statistical methods for the detection of data fabrication in clinical trials. BMJ, 331(7511), 267–143. Yamamoto, K., & Lennon, M. L. (2018). Understanding and Detecting Data Fabrication in Large-Scale Assessments. Quality Assurance in Education, 26(2), 196–212. Prerequisites: General Biology II, Calculus II Biostatistics II --This succeeding course in the sequence will have more emphasis on incorporating journal articles and real world experiments. --Students will have to orchestrate inquisitions by exploratory data analysis and statistical methods involving R. There will be assigned data sets and journal articles to do just that. --This is not a “pen and paper course”. Texts and journal articles will cater to subjects both from prerequisite and this course. Means of data retrieval and manipulation are crucial; it may be the case that the data desired in inaccessible, hence students will have to resort to alternative data sources that yields much different conclusions. NOTE: personally refresh your knowledge and acquired R skills from calculus and Biostatistics I alongside ordained reacquaintances in course. NOTE: this course serves only to towards the perspective of students in the biological sciences, so no one in the biological sciences should be looking elsewhere. MIND YOUR DAMN BUSINESS, AND KEEP YOUR DAMN BUSINESS . Assessment --> Assignment Sets (prerequisite & current level from multiple sources) 15% Analytical and R based 3 Exams (prerequisite & current level) 30% Labs + Data Analysis Term Project 40% 2 Field Inquisitions with R 15% Conducted Journal Articles Computational Inquisitions Supporting data sets to be provided Gov’t administration field experiments inquisitions Assignment Sets --> Will be reacquainted with prerequisite tasks, prerequisite projects AND current level tasks (analytical and R computational). Exams --> Exams will have the same manner of administration and activities as exams from prerequisites. Yet, consisting of both prerequisite tasks AND current level tasks. Limited open notes. LABS WITH R --> Data Processing Data Assimilation (ecological & biological), file types, APIs, etc. Data Wrangling Descriptive Statistics Hypothesis Testing (advance practice from prerequisite) Expt. Design, Multiple Comparisons Regression (Mult. Reg. and Dummy) ANCOVA MANCOVA Non-parametric Statistics (advance practice from prerequisite) Clustering PCA and Kernel PCA Comparing & Averaging Models Analysis of Trait Evolution Fitting models of Trait Evolution TERM PROJECT --> The term project has been broken down into multiple components due throughout the semester to provide further guidance for students. On given date, students will select a dataset to use for their term project. Students can either provide their own dataset (if they have collected data during their research), or will be given the opportunity to analyse a complex dataset supplied by faculty as their term project. For the Hypothesis Activity (given due data), students will take a close look at their dataset and formulate biological hypotheses that they would like to test statistically. The assignment will be handing in these hypotheses. For the Experimental Design assignment (given due), students will outline which analyses they will use to test their biological hypotheses and provide the specific explicit statistical hypotheses that they will test. The Term Project Report (given due having additional 1 week collection buffer) will be written after students complete their analyses. The report will include a Statistical Methods and a Results section, complete with tables and figures. Methods should include sufficient detail to redo the analyses. The results should include everything necessary for interpretation of their analyses and data, but not superfluous material. Term Project Reports for all students should include a title page with a title, student name, course number and name, and assignment name. The text of the report should be double spaced, with indented paragraphs, 1” margins, 12pt Times New Roman Font, and page numbers. Tables should be single spaced with headings above each table. Figures should have captions below each figure. Figures and tables can be embedded in the text or provided at the end of the document. Literature cited should follow the format for the journal Evolution. Assignments that do not follow these formatting instructions will be returned to the student for correction prior to grading. NOTE: most labs done serve as structure for your HA and ED NOTE: I will be collecting your R development (having sensible commentary) for the term project in PDF along with the term report in PDF. Finally, students will give a short, in-class presentation about their study, analyses and findings. Presentations will be in PowerPoint. MAJOR COURSE TOPICS --> Methods of data acquisition, data wrangling and summary statistics (prerequisite reinforcement) Goodness of Fit (prerequisite reinforcement) MLE and MoM Hypothesis Testing (prerequisite reinforcement) Experimental Design & Sampling Regression (multivariate) OLS review (variable selection, summary statistics, forecasting) Quantile Regression compared to Least Squares regression Variable selection, summary statistics, forecasting ANCOVA MANCOVA Non-parametric Statistics Resampling Techniques Clustering (K-means or DBSCAN?) Principal Component Analysis (PCA) and Kernel PCA Model Selection & Likelihood (emphasis on computational logistics and implementation) Phylogenetic Regression Extensions of Phylogenetic Statistics Prerequisite: Biostatistics I Advanced Statistical Modelling and Machine Learning for Biostatistics This course explores advanced statistical modeling techniques and machine learning methods as applied to biostatistical problems. Topics include generalized linear models, hierarchical modeling, Bayesian statistics, and the integration of machine learning algorithms for analyzing complex biological and health data. Note: 2 lectures per week, with approximately 2 hours per lecture. Assignments -- Assignments will be quite laborious in the interest of sustainability with knowledge and skills through your journey in biostatistics. Each assignment will comprise of the following elements: A. problems and tasks encountered in both Biostatistics I & II. Such problems and tasks will also make extensive use of R. As well, being advanced biostatistics students, projects from Biostatistics I & II can/will also be considered basic assignments as well. B. Course level assignments to such given course topics. Good emphasis on ability to comprehend and specify the transition from prerequisite skills/tools to course level tools/method/skills; then implementation. Will also make extensive use of R. Data Science Basics Quizzes -- For the Data Science Basics module there will be handwritten quizzes to test knowledge, comprehension, appropriateness and T/F. Exams -- Exams will account for all modules. Assignments will be strong foresight of what’s to appear on exams. You will be making extensive use of R with open notes for all course modules. Exams will feel like projects where each “project” will involve multiple modules. Make-up Student Project -- Applying Advanced Models to Biostatistical Data. Concerns students who are interested in making up lost weight towards their final grade; the better you did in this course, the lower the value. Can regain up to 5% for final grade. Students will be given a sack to randomly (and blindly) pick a project. Students will have until 2 days before the final grade submission deadline to submit projects. Students will be privately given project details via student email where they will have to acquire the data from specified sources. Course Assessment -- Assignments 20% 3 Exams for all modules 60% 3 Data Science Basics Quizzes 15% Will be precursors to exam(s) for the Data Science Basics module Make-up Student Project (being conditional) COURSE OUTLINE -- WEEK 1-3. Introduction to Generalised Linear Models (GLMs) with model estimation and summary statistics Multilinear Regression (fast fast review) Quantile Regression (fast review) Logistic Regression Poisson Regression WEEK 4-5. Hierarchical Modelling (HM) Introduction to HM Multilevel Modelling Random Effects and Mixed Models WEEK 6-7. Bayesian Statistics in Biostatics Note: I don’t introduce things to be a disgusting, miserable, viral bastard. Module will be extremely goal oriented, namely, problem, goal(s), methodology, logistics, implementation, evaluation. No social nor psychological probes/inquisitions upon students; there are certified licensed professionals elsewhere tied to meaningful or economic interests. Bayesian Inference (to the point, constructive and economical) Markov Chain Monte Carlo (MCMC) Methods – only constructive and economical methods Bayesian Regression Models WEEK 8-9. Advanced GLMs and Extensions Negative Binomial Regression Zero-Inflated Models Generalised Estimating Equations (GEE) WEEK 10-15. Data Science Basics Note: subjects of overfitting or underfitting arise in model validation statistics. Data Acquisition. Data Probing: glimpse(), str() Data Wrangling (functions from dplyr R package with piping) Summary Statistics, Skew, Kurtosis, Correlation Analysis and Heatmaps Machine Learning Overview Feature Selection with R (underlying methods may not be fully comprehended, but that’s generally the world): Principal Component Analysis (PCA), Kernel PCA, Boruta, FSelectorRccp. Comparative observation among such prior three also expected. Multiple Regression (very rapid review) OLS and Quantile Classification Logistic Regression Support Vector Machines Decision Trees Random Forests Clustering (K-Means and DBSCAN advanced repetition) Includes the Elboew Method, Silhouette Score and Davies-Bouldin Index Prerequisites: General Biology II, General Chemistry II, Biostatistics II General Botany The main objective of this course is to introduce the student, who is majoring in Biology, to the wonders of the Plant Kingdom and its (often) closely allied Kingdoms of Bacteria, Archaea, Fungi, and Protista. Applied Botany will be a focal point of the laboratory exercises in hopes of creating an ongoing interest in the science of plants. Literature: Mauseth, J. D. (2019). Botany: An Introduction to Plant Biology. Jones & Bartlett Learning Lab Manual: Mauseth, J. D. (2019). Botany: A Lab Manual. Jones & Bartlett Learning Assessment: 3 Exams Labs Final Exam Labs --> Lab topics will be chosen to coincide with lecture topics where possible. Leaf identification of selected tree species will also be an integral part of lab. Course Outline --> General Introduction The Development of Plant Study/What is Plant Biology? Differentiation between plantae, Protista and Fungi. Common misconceptions in aquatic environments Animal cell versus the plant cell Plant Cell Structure & Function Plant Metabolism/Photosynthesis Plant Metabolism/Respiration Plant Growth and Development Plant Hormones and Environmental Response EXAM I Plant Tissues Roots Stems Leaves EXAM II Reproductive Morphology/Flowers Reproductive Morphology/Fruit Reproductive Morphology/Seeds The Plant Life Cycle/Meiosis and Alternation of Generation Movement of Water & Solutes in Plants 9 Soils EXAM III Classification & Systematics Bacteria & Viruses Fungi & Lichens Protistans/Algae Bryophytes Ferns & Their Relatives Gymnosperms 22 Angiosperms FINAL (comprehensive) Prerequisites: General Biology I & II, General Chemistry I & II Plant Symbioses Course aims to foster a deeper comprehension of the crucial role symbioses play in shaping the diversity of life on the planet. Devoting roughly equal time to discussing broad concepts – such as generalities among symbioses, origins and establishment of symbioses, coevolution and co-speciation – and learning the specifics of well-studied exemplars of plant-bacterial, plant-fungal, plant-animal, and plant-plant symbioses. Often a 2 credit course. Upon completion of this course, you will be able to: Define symbiosis in several ways Describe the evolutionary and ecological significance of symbioses Explain our current understanding of how symbioses form and how they are maintained Discuss how symbiosis can lead to diversification Summarize the specifics of some symbioses involving Plants and bacteria Plants and fungi Plants and animals You will also practice: Evaluating and synthesizing journal articles Leading a discussion with classmates Literature: The Symbiotic Habit, Angela E. Douglas. 2010. Book excerpts and journal articles Assessment: Quizzes (10%) Attendance + Conduct + Written discussion synopses (30%) 3 Exams (60%) Course Outline Week (1) Introduction to Symbiosis; significance Case: ant plants Week (2) Example symbiosis 2 – Mycorrhizae Defining symbiosisWeek (3) Example symbiosis 3 - Grass endophytes Symbiotic spectra from antagonist to mutualist Week (4) Example symbiosis 3 – Cyanobacteria Evolution and partner capture Week (5) Example symbioses 4 - Pollination symbioses - Ficus and Yucca Costs of symbiosis and cheating Week (6) Vertical transmission and assimilation Example symbiosis 5 - Legumes and rhizobia Week (7) Establishment of symbioses Diversification and coevolution Prerequisites: General Biology I & II, General Chemistry I & II Field Botany This course is designed as an introduction to the diversity of plants found in natural systems. Since it is a field course, most of the students' time will be spent either in the field or working with specimens collected from the field. However, there are some basic ideas about systematics in general, plant systematics specifically, and plant evolution that will be presented in class. In addition, students will demonstrate proficiency with the basic terminology of plant morphology and plant taxonomy. The student will learn to collect plant specimens, preserve them, and present them in a format similar to specimens found in herbaria. Since habitat information will be taken with each specimen, students will learn to associate plants with different habitat types. In addition to the field elements, the student will learn to use the botanical literature and web-based resources needed to identify a specimen. Noteworthy: appropriate apparel and clothing expected. Please don’t insist on wearing full jet-black or heavy dark colours...but avoid white, off-white, cream or butterbean. Eco-friendly bug repellent/sunscreen, drinkable water, etc. Head gear for heat if need be, etc. A handy watch. An insulated smartphone (strictly for GPS, photos and emergencies if need be). Noteworthy: THIS IS A RAIN OR SHINE COURSE Course literature: A scholarly text of flora of the ambiance of concern A wildflower guide text Key events during course --> Initiate collections and begin preservation Examination Complete collections and preservation Complete identification Complete mounting Present collection for grading No collections or specimens will be accepted after due date Noteworthy: students should have decent and competent skills for record of specimen. Data for samples should be augmented with location, date, time, etc., etc. Objectives for Taxonomy vs Systematics --> The student will be able to: -Describe the function of classification -Distinguish between taxonomy and systematics and be able to identify a classification as systematic or taxonomic -Describe the reasons for preferring natural classifications over artificial classifications -Describe the reason that classical taxonomy is an hierarchical scheme of classification -Describe the role that key characteristics play in taxonomy -Describe why consistency is both valuable for taxonomy and hard to achieve. -Distinguish between phenetics and cladistics -Describe the essential features of a dendrogram, including its shape and the assumptions about time inherent in a dendrogram -Relate the reason that botanical taxonomy uses "division", rather than "phylum" as the hierarchical level below that of kingdom and above that of class -Define the following terms: Plant Morphology Classification Taxonomy Systematics Natural Classification Binomial Hierarchy Key Characteristic Consistency Phenetics Cladistics Dendrogram Division Phylum Objectives for Morphology --> The student will be able to: -describe and identify from a living plant the following structures: Stem, Leaf, Bud, Root, Node, Lenticle, Bract, Leaf Parts (Blade, Petiole, Midrib, Stipule), Leaf Veination (parallel, pinnate, reticulate), Leaf Margins (Smooth, Dentate, Serrate, Lobed Crenate), Leaf Arrangements (Alternate, Opposite, Whorled, Simple, Compound, Palmate, Pinnate, Bipinnate), Leaf Shapes (Elliptical, Obovate), Leaf Tip Shapes (Acute, Accuminate), Flower Types (Complete, Incomplete, Perfect, Imperfect), Flower Parts (Receptacle, Perianth, Calyx - Sepal, Corolla - Petal, Stamen, Anther, Filament, Pistil, Stigma, Style, Ovary, Carpel [Simple, Compound]), Flower Shapes (Radial, Bilateral [Zygomorphic], Superior, Inferior), Fruit Types (Drupe, Berry, Hesperidium, Pepo, Pome, Aggregate, Multiple, Samara, Nut, Grain, Achene, Capsule, Silique, Legume, Follicle) Objectives for Plant Kingdom--> The student will be able to: -Describe the common characteristics all plants share -Identify a specimen as a Bryophyte -Describe the characteristics that separate Bryophytes from other required plant divisions (see below for which plant divisions are required). -Identify a specimen as a Equisetophyte -Identify a specimen as a Lycopodiophyte -Identify a specimen as a Pteridiophyte -Describe the differences between Vascular Plants and Non-Vascular Plants. -Describe the differences between Flowering Seed Plants and Non-Flowering Seed Plants. -Identify a specimen as a Coniferophyte -Identify a specimen as a Cycadophyte -Identify a specimen as a Pteridiophyte -Identify a specimen as a Ginkophyte -Describe the differences between Monocots and Dicots in cotyledon number, leaf veination, flower part number, arrangement of stem vascular bundles, presence of secondary growth, and the manner in which roots originate.Identify a specimen as a Magnoliophyte -Identify a specimen as a Liliopsid -Identify a specimen as a Magnoliopsid -Define the following terms: Liana Epiphyte Cone Evergreen Deciduous -Diagram the Dendrogram of the Plant Kingdom presented on the web page Objectives for Plant Family --> For the following plant families (minimum): Aceraceae Cactaceae Compositae Curcubitaceae Fabaceae Fagaceae Orchidaceae Poaceae Rosaceae Solanaceae The student will be able to: Give the common and scientific names for the family; describe the general leaf, flower and fruit characteristics of members of the family; describe the importance of the family to the human economy and the uses to which members of the family have been employed. Note: there are around 24 characters for learning families of flowering plants. Objectives for Collection --> The student will collect, identify, and mount 25 specimens of local plant species. The student will present a collection that includes specimens from at least two plant taxonomic group above that of the family. The student will present a collection that includes specimens from at least ten different plant families. Course Assessment --> Evaluation will be based on a single examination and the plant collection to be done by each student. Each specimen must be presented properly preserved and identified (using the traditional hierarchical scheme of Kingdom, Division, Class, Order. Family, Genus, and Species). In addition, information on the collection date and locale and habitat must also be presented. Only specimens complying with all requirements will count toward the student's total. To learn what constitutes an acceptable specimen, go to the “Preservation”, “Mounting” and “Examples” guides. As the objective of this class is to learn about the local natural flora, horticultural cultivars (including hybrids) will not be accepted as specimens. In addition, the collection must present specimens from at least 10 different plant families and from at least two different taxa above the level of Family (i. e., from at least two different Classes or Orders). Failure to satisfy these criteria will result in the loss of particular points for the former and particular for the latter, subtracted from the total points earned. Once they are graded, the portfolios are the property of the student and can be retrieved from the instructor. They will be discarded on designated date if not picked up before. Class Attendance 5% Quizzes 15% 2 Exams 30% Collection (quantity, order, accuracy, quality) 50% NOTE: for class attendance there will be a threshold for days missed towards failure of course. For conduct in the field and in lectures there will be a policy towards pass of fail; includes, littering, pollution, texting, social media, music in the field, habitat damage, savagery upon living creatures. Prerequisites: General Biology I & II, General Botany Plant Physiology This course is designed to provide students with comprehensive exposure to the subject of plant physiology. The laboratory exercises provide hands-on experiences with experiments and training in instrumental skills. Topics include: water relations, photosynthesis, inorganic nutrition, metabolism of organic materials, and plant growth regulation, with emphasis on environmental factors in the physiology of plants. Lecturing Text --> Introduction to Plant Physiology, 3rd. William G. Hopkins Other references: Plant Physiology, 2nd. L. Taitz and E. Zeiger Plant Physiology, 4th. F. Salisbury and Cleon W. Ross Resources to accompany designated lab manual --> PHWYE – Plant Physiology/Botany: https://www.phywe.com/en/biology/university/plant-physiology-botany/ Journal of Experimental Botany (https://academic.oup.com/jxb) Park, S. (2021). Plant Tissue Culture: Techniques and Experiments. Academic Press By the end of this course, the student will be able to --> 1. Comprehend the fundamental concepts of plant physiology 2. Describe the physiological mechanisms of plant growth, function, and development 3. Recognize and describe how plants respond to their environment Assessment --> Quizzes 10% 3 Exams 60% Laboratory 30% Activities Reports and Presentations Lab reports and presentations --> Laboratory work is done in small groups. To foster learning and interaction among students, each group will design an experiment to conduct based on techniques learned in the course. Learning goals assessment--> Specific questions on exams and participation in class will be used to assess student knowledge of course learning goals, including demonstrated mastery of fundamental terms and mechanisms in plant physiology. In graded laboratory exercises, students will communicate their understanding of techniques used in the discipline. COURSE OUTLINE --> Introduction Course overview: the organization of plants and plant cells Part I: Water and mineral nutrients Water in plant cells Water relations of the whole plant Essential nutrients Nutrient uptake Exam I Part II: Photosynthesis and Respiration Photosynthesis: light and pigments Photosynthesis: light reaction Photosynthesis: carbon assimilation Photosynthesis: carbon allocation Respiration Exam II Part III: Regulation of plant growth and development Cellular basis of growth and development Plant hormones Auxin 16 Gibberellins Cytokinins Abscisic acid and ethylene Photomorphogenesis: responding to light Plant movements Photoperiodism Temperature control Part IV: Stress physiology and biotechnology Plant response to environmental stresses Biotechnology Exam III LABS --> Week 3 -- Lab 1 Water Relations Time domain reflectometry Relative water content Osmotic adjustment Transpiration rate Lab 2 How do plants/trees transport water under negative pressure? Replicate experimentation to best of ability and speculate/identify elements in plant/tree physiology that make transportation with negative pressure possible Shi, W., Dalrymple, R.M., McKenny, C.J. et al. Passive water ascent in a tall, scalable synthetic tree. Sci Rep 10, 230 (2020). If such process is innate in trees/plants can they function well in alien environments such as space, Moon’s surface, or environements with larger gravitation? Week 4 -- Lab 3 Nutrition Deficiency symptoms Nitrate-nitrogen concentration Week 5 -- Lab 4 (Part 1) Photosynthesis Photochemical efficiency Stomatal characteristics Leaf area meter Week 6 -- Lab 5 (Part 2) Photosynthesis Chlorophyll content Light meter Net photosynthetic rate Week 7 -- Lab 6 Respiration Net respiration rates Temperature and respiration Starch quantification Week 9 -- Lab 7 Plant Growth Regulators Effects of gibberellic acid on germination and growth Week 10 -- Lab 8 Plant Growth Regulators Lab 6 (cont.) Effects of auxin on rooting Week 11 -- Lab 9 Light Response Quantification Intensity/duration Direction Week 13 -- Lab 10 Abiotic Stress Drought Salinity Flooding Week 14 -- Wrap Up Prerequisites: General Botany, Organic Chemistry I and/or Biochemistry I Plant Systematics Will explore the origin and diversification of land plants while emphasizing flowering plants. You will become familiar with --Taxonomy (identification, nomenclature, classification emphasizing flowering plants), --Evolution (speciation, reproductive biology, adaptation, convergence, biogeography) --Phylogenetics (phenetics, cladistics, morphology and molecules). Labs emphasize learning representative families and genera of flowering plants in your ambiance with use of keys and manuals. A plant collection of 25 species is done. Assessment --> Labs + Lab quizzes Field Trips + Field Trip quizzes 2 Exams Texts --> Clark et al. 2014. Plant Systematics: Laboratory Manual and Supplementary Resources Judd, W. S. et al. 2008. Plant Systematics: A PhyloGenetic Approach. Sinauer Inc. Resources --> Manual for recognising the (50) most common plant families List of common plant families and major plant groups you should learn to identify Morphology vocabulary manual, and a list of plant morphology words you should know to help you identify and describe plants. Tools --> Hand lens (10X; preferably with neckband so you don’t drop it and loose it Digital camera (smartphone or tablet with camera is OK) - you will not be able to participate in class if you do not have access to a digital camera from which you can download photos Medium size notepad and lots of writing utensils; all should be sealable in a Ziplock Computer or tablet with internet access Come prepared with the following --> - Field boots or shoes (good ankle support is important, and there are rattlesnakes out there) - Hand lens - Ambiance flora (for keying practice) - Hat - Sunscreen and lip protectant - Pocket knife if you have one - Ziplock bags/containers/bags for collecting - Rain gear (if it’s threatening) - Insect repellent (eco-friendly) - Competent hydration fluid Laboratory--> Many of the laboratory sessions will be devoted to field trips. Other lab sessions will consist of a directed overview of the plant groups covered in the lecture. You can also work on identifying your plant collections during lab time. Attendance at lab is required. Most labs will have a quiz reviewing material from the lab. Periodically we will assign homework plants. These will consist of unknown plants that you attempt to key out with your textbooks. You will do this on your own time. Each lab quiz or homework will be worth 10 points, with a total of 100 lab quiz points for the semester. If we have more than a total of 100 possible lab quiz points, you may drop your lowest scores and count only the 10 best quiz grades Field Trips --> Several field trips have been scheduled during laboratory time. Attendance is required. Sometimes we may be a little late in returning. Activities take place whether rain or shine. you will collect and identify 25 plants that are in flower and/or fruit and make botanically accurate labels to accompany your specimens that note the collecting locality, date, collector, and additional attributes of the plant. More details will be forthcoming on this exercise. Quizzes and Tests --> Each field trip will include a quiz. Most of these will ask you to identify plants we have learned. Some may require you to key out “unknowns” with the help of your texts. We will also periodically hand out homework that should be done individually and out of class. We will also devote part of one lecture each week to quizzes to review your knowledge of plant families. Exams will cover lecture and lab material. Part of each exam will be a “practicum” asking you to identify various plants or answer questions about them. Prerequisite: Field Botany Dendrology Course serves to provide the student with an intensive and broad experience with respect to knowledge of trees for majors in botany. This experience will include identification and distribution of native and introduced trees, their biological characteristics, identification, habitat and ecology. Goals --> Describe important aspects of the morphology, anatomy, development and reproduction of trees using correct terminology. Describe the structure, function and importance of modern nomenclature and taxonomic systems. Describe the morphological, geographic, ecological and economic characteristics of important forest tree species of region or ambiance. Explain basic population, community ecology, and ecosystem-level concepts as it relates to the Plant Kingdom. Evaluate ecology and diversity in a global context and specifically in whatever region Demonstrate the ability to identify and classify trees and be able to problem solve and identify unknown species. Noteworthy: appropriate apparel and clothing expected. Please don’t insist on wearing full jet-black or heavy dark colours...but avoid white, off-white, cream or butterbean. Eco-friendly bug repellent/sunscreen, drinkable water, etc. Head gear for heat if need be, etc. A handy watch. An insulated smartphone (strictly for GPS, photos and emergencies if need be). Noteworthy: students should have decent and competent skills for record of specimen. Data for samples should be augmented with location, date, time, etc., etc. NOTE: THIS IS A RAIN OR SHINE COURSE Etiquette and formality in assessment for tools, analysis and data collection (procedures and rules) stemming from field activities will be given to students concerning the following attributes: Accuracy and location Quality Quantity Organisation NOTE: unique portfolio for mangroves, seashore trees and fruit bearing trees. Literature --> A Text of Dendrology Regional Dendrology resources (long range in time) Course Assessment Quizzes --> Family, Species, Genus, Common Name, geography, habitat, ecology Midterm Final Portfolio Presentation Prerequisites: General Biology I & II, General Botany, Plant Physiology. Aquatic Botany Course objectives: Aquatic plants are generally defined as those higher (vascular) plants completing their life cycles wholly or partly in a submerged state or in saturated soil. As well, course will treat algae due to similar functions as plants in the ecosystem. The goals of this course are: i) to learn the basic taxonomy of common aquatic plants and algae ii) to become familiar with the habitats where aquatic plants and algae are commonly found, iii) to understand the functioning of nutrient cycles in aquatic systems iv) to know the various definitions of wetlands, marine forests and important legislation applicable to wetlands and marine forests v) to understand the concepts of mitigation, restoration, constructed wetlands, marine forests, effluent dominated streams and wetlands, and how these are implemented vi) become familiar with control and management of aquatic plants and algae in perturbed and man-made ecosystems vii) become familiar with aquatic nuisance plant/algae species and their role in the environment viii) become familiar with the primary literature (scientific journals and reference books) in this field. The lab portion will focus on use of small ecosystems for study, short field trips to local wetlands, marine environments and familiarization with field instruments and water testing kits. NOTE: course has a rain or shine policy with any outdoor activity Literature (all necessary)--> Comprehensive texts on wetlands emphasizing, taxonomy and ecology Comprehensive texts on aquatic/marine algae emphasizing, taxonomy and ecology Comprehensive texts on the physiology and regulation of aquatic/marine plants and algae Journal articles Credible scientific agencies/ministries Exams --> Exams will be comprehensive and questions will come from lectures, textbooks, labs, student presentations, handouts and field trips. (Hint: topics which arise in two or more of these areas are the most likely to show up on exams.) Mid-terms will be reviewed in the class period following the exam. Identification and Collection --> Identification will reinforce professional scientific standards in taxonomy, vocabulary and data structure. Collection ability dependent on government policy with respective ecosystem. Labs + Field Activities --> The lab portion will focus on use of small ecosystems for study, Various short field trips to local wetlands, marine environments Familiarization with field instruments and water testing kits Under the Scope: microscopy techniques to visualize anatomies & measure structures; techniques may have some disparities to the usual micrscopy activity concerning aquatic plants, algae Photosynthesis lab with aquatic plants and algae Group Term Paper --> Term paper to be a review of a topic of interest within the fields of aquatic plants or algae or wetlands/aquatic/marine biology or ecology. Will be presented to the class. Scientific format and references (texts, journal articles and credible scientific agencies/ministries) are expected. Group Research Project --> Design, conduct and report a field or lab experiment developed with the instructors. Assessment --> 2 mid-terms 30% Final 15% Papers 20% Labs and plant/algae collection 20% Presentation & participation 15% Course Outline --> Definitions and Wetlands, Aquatic and Marine ecology/habitats Wetlands of North America Classifications, Inventory and Delineations Inland Wetlands Salt Marshes Field Trip to Herbarium Mangroves Sea plants/trees and algae Field Trip(s) to Marine ecosystems Hydrology, Water Budgets and Models Nutrient Cycling Wetlands/Aquatic/Marine Biological Adaptations Limnology Riparian Zones and Regional Wetlands Characterisation, Demography, Human interaction Sea Grass Beds Algae Field Trips to regional wetlands Riparian Zones Rivers of the region Seasonal wetlands Invasive Species - Aquatic Nuisance Species (impacts and laws) Animalia, plantae and algae Wetland Management, Laws, Protection Marine Management, Laws, Protection Control & Management of Aquatic Plants with Physical & Chemical- Handouts Control & Management of Aquatic Plants - Handouts Mechanical & Biological Controls Environmental Issues - Case Studies Student Presentations Student Presentations Prerequisites: General Botany, Plant Physiology, Analytical Chemistry Organic Chemistry I In-depth study of: (i) the structure of organic compounds and the functional groups (bonding, acid-base properties, nomenclature, conformations, stereochemistry), and (ii) the synthesis and reactivity (including detailed mechanisms) of alkanes, alkenes, alkynes, halides, alcohols, ethers, epoxides, sulfides and organometallic reagents. Laboratory experiments are related to topics covered in lecture and emphasize organic laboratory techniques, synthesis and spectroscopic characterization of organic molecules. Typical Texts: McMurry, John E. Organic Chemistry. 8th Edition. Brooks/Cole, 2012. McMurry, Susan. Study Guide with Student Solutions Manual. 8th Edition. Brooks/Cole. Typical Lab Manual: Barbaro, John and Richard K. Hill. Experiments in Organic Chemistry. 3rd Edition, Contemporary Publishing Company of Raleigh, Inc., 2006 Grading: 3 Exams (50% combined) Cumulative Final Exam (25%) Labs (25%) (On the occasion of significant improvement on the final exam, more weight will be placed on the final exam) INSTRUCTIONAL METHODS: List the different instructional methods you might use, in the course of the semester. List supplementary learning options, if any: Traditional lecture with use of chalkboard Computer assisted diagrams and graphics Molecular Models Team work in the laboratory Homework assignments Solving specific questions related to content studied Written exams and distribution of study questions/previous exams Use of the Internet UNIQUE ASPECTS OF COURSE (such as equipment, specified software, space requirements, etc.): Organic chemistry laboratories and their associated equipment, instruments and chemicals. Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Ch. 1 Structure and Bonding Bonding; Hybridization; Drawing Chemical Structures; Functional Groups; Intro to IR Spectroscopy Ch. 2 Polar Covalent Bonds; Acids and Bases Chemical Bonding (Ionic and Covalent); Electronegativity and Dipole Moments; Formal Charges; Resonance Structures; Acid Base Theory (Bronsted-Lowry, Lewis); Acid and Base Strength (pKa); Acid-Base Reactions; Organic Acids and Organic Bases Ch. 3 Organic Compounds: Alkanes and their Stereochemistry Alkanes, Alkane Isomers, and Alkyl Groups; Properties of Alkanes; Conformations Ch. 4 Organic Compounds: Cycloalkanes and their Stereochemistry Cis-Trans Isomerism in Cycloalkanes; Stability and Conformations of Cycloalkanes; Chairs Ch. 5 Stereochemistry at Tetrahedral Centres Enantiomers, the Tetrahedral Carbon and Chirality; Optical Activity; R/S Sequence Rules; Diastereomers and Meso Compounds; Racemic Mixtures, Resolution of Enantiomers; Prochirality; Chirality in Nature Ch. 6 An Overview of Organic Reactions Kinds of Organic Reactions (Radical and Polar); Mechanisms; Describing a Reaction (Equilibria, Rates, Energy Changes, Bond Energy; Transition States, and Intermediates) Ch. 7 Alkenes: Structure and Reactivity Preparation and use of Alkenes; Cis-Trans Isomerism; Alkene Stereochemistry and E/Z Designation; Stability of Alkenes; Electrophilic Addition Reactions; Markovnikov’s Rule: Carbocation Structure and Stability; Carbocation Rearrangements Ch. 8 Alkenes: Reactions and Synthesis Preparation of Alkenes via Elimination Reactions; Addition Reactions of Alkenes (Halogenation, Hydration, Halohydrins, and Hydrogenation); Oxidation of Alkenes (Epoxidation and Hydroxylation); Addition of Carbenes; Radical Additions to Alkenes (Polymer Formation); Reaction Stereochemistry Ch. 9 Alkynes: An Introduction to Organic Synthesis Preparation of Alkynes; Addition Reactions of Alkynes (X2, HX, H2O, H2); Oxidative Cleavage; Alkyne Acidity and Alkylation; Introduction to Organic Synthesis Ch. 11 Reactions of Alkyl Halides: Nucleophilic Substitutions and Eliminations SN2, SN1, E2, E1, E1cB Reactions; Zaitsev’s Rule; Deuterium Isotope Effect Ch. 10 Organohalides Preparation of Alkyl Halides and Grignards; Radical and Allylic Halogenation; Organic Coupling Reactions, Redox in Organic Chemistry Ch. 17 Alcohols and Phenols Properties of Alcohols and Phenols; Preparation and Reactions of Alcohols; Reactions of Phenols Ch. 18 Ethers and Epoxides; Thiols and Sulfides Synthesis and Reactions of Ethers; Cyclic Ethers (Epoxides); Reactions of Epoxides: Crown Ethers; Thiols and Sulfides LABS --> Some experiments require more than one lab period to complete. Based on an instructor’s preference, availability of equipment/supplies or constraints within a given semester, this laboratory schedule is subject to change, including but not limited to, the addition or replacement of one or more of the above experiments with the following experiments: Addition of Bromine to E-Cinnamic Acid in Methylene Chloride Substitution Reactions of Alkyl Halides: Relative Rates Triphenylmethanol with Hydroiodic Acid 1. Check-in, Laboratory Safety, Practices and Waste Disposal. Simple Distillation. 2. Spectroscopy: Introduction to Infrared Spectroscopy. 3. Recrystallization, IR and Melting Point of benzoic acid. 4. Extraction of Organic Compounds from Natural Sources: Trimyristin from Nutmeg. 5. Paper Chromatography 6. Dehydration of Cyclohexanol. 7. Dimerization of 2-Methylpropene 8. Preparation of Diphenylacetylene Starting from Trans-Stilbene. 9. Preparation of Butyl Bromide/Preparation of t-Butyl Chloride (SN2/SN1). 10. Oxidation of Isoborneol to Camphor. 11. The Williamson Ether Synthesis: Preparation of Aryloxyacetic Acid from Cresol. Prerequisites: General Chemistry II Biochemistry: The study of biochemistry investigates the interplay between biological macromolecules such as proteins and nucleic acids, and low molecular weight metabolites (such as the products of glucose metabolism). In this course, you will apply your knowledge of intermolecular forces, thermodynamics (when a reaction occurs), chemical kinetics (how fast a reaction occurs), and chemical structure and functionality to understand how biological molecules (and life) work. COURSE GOALS AND OBJECTIVES (Our Roadmap!) -Be able to describe/identify the forces that direct/stabilize different levels of protein structure -Be able to predict how changes in amino acid (or nucleotide) sequence can affect macromolecular structure and function -Be able to explain how enzymes are able to affect reaction rate enhancement -Be able to articulate and apply what the enzyme parameters of KM, Vmax, kcat and kcat/KM tell us about an enzyme -Be able to describe the interactions of biomolecules both quantitatively and qualitatively (in many cases, including mechanistic details) -Be able to understand the flow of metabolic intermediates through a pathway and communicate information about metabolic pathways using diagrams -Be able to describe multiple experimental methods used in biochemistry, interpret data from these methods to form conclusions, and develop a testable hypothesis to answer a question -Be able to summarize and analyse primary literature and data, and apply gathered information to new situations -Increase problem solving skills such as: critical thinking, data analysis, graphical analysis -Increase process skills such as: communication of scientific concepts and experimental results, group dynamics and teamwork, management and self-assessment -Develop a community of active learners who are intentional about their educational choices Course Materials: Calculator Emphasis on reinforcing skills with software --> << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Typical Texts --> Nelson DL and MM Cox. Lehninger Principles of Biochemistry (5th edition). (“Lehninger”) Loertscher J and V Minderhout. Foundations of Biochemistry (3rd edition). (“FOBC”) Additional --> Blast protein databases, align protein sequences, build protein homology models, and evaluate the quality of these models Lab Manual example --> Lasseter, B. F. (2020). Biochemistry in the Lab: A Manual for Undergraduates. CRC Press Related to week 6: Edwards, P., Zhang, C., Zhang, B. et al. Smartphone based optical spectrometer for diffusive reflectance spectroscopic measurement of hemoglobin. Sci Rep 7, 12224 (2017). Course Overview --> You will frequently be given initial assignments to work on as an individual before class. These assignments must be ready at the start of class – your preparation will form part of your weekly participation grade. During our class meeting time, you will frequently function as a member of a Learning Team, developing and examining chemistry concepts as a unit. Your team effort and participation is part of your weekly participation grade. The team responses to a few Key Questions on each in-class activity will be evaluated for strength of concept and effective communication of the concept. The team will also strategize on ways to improve teamwork and team products. These responses will also form part of your weekly participation grade. Application exercises will be assigned for each activity. Together with problems from the text, they will form your weekly problem set that will be collected and graded for each individual. These homework problems and exercises are important to your success in the course. Actively working these homework problems is essential for your understanding of the material, as they bring your concept development full circle. The questions will be drawn from lectures, in-class activities, problem sets and discussions, as well as relevant primary literature that you may not have been previously assigned. The purpose of doing biochemistry is to gain experience in experimental methods that you’ll be reading about throughout the semester. Attendance on your scheduled lab day is expected. Software activities concerning biochemistry will accompany labs. Software activities concerning biochemistry will accompany labs as pre-lab development or post simulations. Grading --> Team Participation Problem Sets/Other Laboratory 2 Midterm Exams Final Exam Lecture Outline --> Week 1 Introduction to Biochemistry Week 2 Intermolecular forces and water. Amino acids and peptide bonds Week 3 Protein Folding Week 4 Working with proteins Week 5 Enzyme catalysis. Enzyme Kinetics Week 6 Enzyme inhibition. Hemoglobin Week 7 Exam 1; Carbohydrates Week 8 Glycobiology Week 9 Lipids and membranes. Transport across membranes Week 10 Signal transduction. Metabolism overview Week 11 Glycolysis. Glycolysis regulation and related pathways Week 12 Glycogen metabolism and gluconeogenesis. Citric Acid Cycle Week 13 Electron Transport Chain / Oxidative Phosphorylation; Exam 2 Week 14 Lipid metabolism. Nucleotides and nucleic acids Week 15 Nucleic acids structure and function Week 16 Final Exam Prereqs: General Biology I. Co-requisite or Prerequisite: Organic Chemistry I Plant Nutrition Fundamentals of plant nutrient availability, uptake, assimilation, transport, function, and deficiencies. Influence of plant root environment and root physiology on plant nutrient status and subsequent effect on plant growth, crop yield, and relationship to plant diseases and pests. Texts of interest: Mengel, Konrad and Ernest a. Kirkby (eds). Principles of Plant Nutrition. Fifth edition. Dordrecht: Kluwer Academic Publishers, 2001, 849 pages 2, 651 pages Marschner, Petra( ed). Marschner’s Mineral Nutrition of Higher Plants. Amsterdam: Elselvier, Labs --> Course will have administered labs based on the following: Motsara, M. R. & Roy, R. N. (2008). Guide to Laboratory Establishment for Plant Nutrient Analysis, FAO Fertilizer and Plant Nutrition Bulletin 19 https://www.ndsu.edu/pubweb/chiwonlee/plsc211/labmanual/lab5.PDF http://schulte.faculty.unlv.edu/BIO442/Lab4Mineral.pdf https://www.globe.gov/documents/355050/41927208/BasicExperiments_MineralNutrition.pdf/85690c28-3497-4729-a659-b1e9c6230ab3 NOTE: if there is any need of Analytical Chemistry lab techniques, they will be done as a precursor to relevant chosen plant nutrition labs. Quizzes --> There will be 3 – 5 quizzes throughout the course Exams --> There will 4 exams for students partake in. There is no cumulative final exam. Assessment: 3-5 Quizzes 10% Labs 30% 4 Exams 60% Course Outline: Week (1) Introduction to Plant Nutrition (Chapter 1) The Soil as a Plant Root Medium (Chapter 17 pp. 418‐424) Nutrient Availability (Chapter 12, Chapter 11 pp. 299‐300) Week (2) The Root (Chapter 13) The Rhizosphere (Chapters 14 and 15) Week (3) Nutrient Uptake (Chapter 2) Water Relations & Long Distance Transport in the Xylem & Phloem (Ch 3) Week (4) Uptake and Release of Nutrients by Aerial Plant Parts (Chapter 4) Photosynthesis and Assimilation of Carbon Dioxide (Chapter 5 pp. 85‐95) Week (5) Yield and Source‐Sink Relationships (Chapter 5 pp. 95‐133) Week (6) Nitrogen (Chapter 6 pp. 135‐142, Chapter 16) Nitrogen (continued) (Chapter 6 pp. 142‐151) Week (7) Sulphur (Chapter 6 pp. 151‐158) Phosphorus (Chapter 6 pp. 158‐165) Week (8) Potassium (Chapter 6 pp. 178‐189) Calcium (Chapter 6 pp. 171‐178) Week (9) Magnesium (Chapter 6 pp. 165‐171) Week (10) Iron (Chapter 7 pp. 191‐200) Manganese (Chapter 7 pp. 200‐205) Week (11) Zinc (Chapter 7 pp. 212‐223) Copper (Chapter 7 pp. 206‐212) Molybdenum (Chapter 7 pp. 226‐233) Week (12) Boron (Chapter 7 pp. 233‐243) Nickel (Chapter 7 pp. 223‐226) Chlorine (Chapter 7 pp. 243‐248) Week (13) Beneficial Nutrients – Sodium, Silicon, and Cobalt (Chapter 8 pp. 249‐263) Week (14) Diagnosis of Nutrient Deficiencies and Toxicities (Chapter 11) Nutrition and Plant Quality (Chapter 9) Week (15) Plant Nutritional Status and Plant Diseases and Pests (Chapter 10) Prereqs: Calculus II, General Botany, Plant Physiology, Organic Chemistry I, Biochemistry I Plant Biochemistry Topics are taught in the context of plant biology. Successful completion of this course will provide students with fundamental knowledge of biochemistry and specific knowledge of compounds and biochemical pathways that occur in plants. Topics include 1. The biochemistry of amino acids and proteins, sugars and carbohydrates, and lipids. 2. Biochemical processes and metabolic pathways specific to plants, including photosynthesis, photorespiration, cell wall biosynthesis, nitrate and sulfate assimilation, distinctive aspects of central metabolism, and plant secondary metabolism. 3. Metabolism in a structure-function context from molecular to subcellular, cellular, organ, and whole-plant levels. 4. Quantitative aspects of biochemistry including enzyme kinetics, protein ligand binding, analytical techniques, and bioenergetics. Learning Objectives and Outcomes Students will • understand plant cell structure, organization, and apply specific biochemical functions to all compartments of the plant cell. • learn the structure, function and biosynthetic pathways of essential biochemical molecules including their key chemical and physical properties. • learn amino acid structures and relate their chemical properties to the synthesis and function of proteins and enzymes. • understand protein structural hierarchy and relate structure to function. • understand how light energy is captured and used to provide chemical forms of energy to power the functions of cells and whole plants. The importance of CO2 fixation and carbohydrate metabolism will be presented. The nature and composition of plant cell walls will be explored. • understand central metabolism, its plant-specific components, and their functional significance at multiple levels. • learn about the rich diversity of secondary compounds and metabolism in plants and how such compounds contribute to human health. • learn principles of enzyme kinetics and apply these through hands on problem sets. Students will be shown how enzyme properties contribute to metabolic processes. • explore principles of metabolic modelling. Required Textbooks 1. Biochemistry & Molecular Biology of Plants, Second edition, print or electronic version, 2015, Wiley Blackwell 2. A general biochemistry textbook Supporting Textbook Gleason, Florence K. (2012) Plant Biochemistry Lab Guide --> J. A. Bryant, P. M. Dey, Jeffrey Barry Harborne (1993). Methods in Plant Biochemistry. Academic Press Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well to this course that further encourages a modern and profession environment, extending beyond memor based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities on e is interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Additional --> Blast protein databases, align protein sequences, build protein homology models, and evaluate the quality of these models Assessment --> Homework and Quizzes 10% 5 Exams 50% Labs 40% Labs --> Software activities concerning plant biochemistry will accompany labs. Software activities concerning plant biochemistry will accompany labs as pre-lab development or post simulations. Labs (active) will have focus on Plantae Course Outline --> Plant cell structure and compartments Amino Acids Structure and properties Ionization and titration Peptides, Properties and purification methods Protein Purification Structure (example: Rubisco) Enzymes and catalysis Enzyme Structure/Function relationships Rubisco Function Oxidation/reduction, bioenergetics, ATP and NAD(P)H Photosynthesis Light absorption Electron Transport Q-cycle and ATP synthesis Bioenergetics, ATP and phosphorylation Sugar structure and function Calvin Cycle Rubisco; photorespiration C4 Metabolism, CAM Metabolism Sucrose: synthesis, transport, breakdown, signals Polysaccharides Starch structure, metabolism Cell wall structure, metabolism Glycolysis Mitochondrial functions Citric acid cycle Electron transport Other Oxidative pentose phosphate pathway Regulation of primary metabolism Nitrogen Metabolism: Fixation Nitrogen Metabolism: Assimilation and GS/GOGAT Sulphur Metabolism: Assimilation and impacts Fatty acid Structure Desaturation Synthesis I & II Oxidation I & II Health promoting secondary products Flavonoids I & II Phenolics and ESPS synthase Terpene synthesis Carotenoids Alkaloids I & II Protein-Ligand Interaction I – III Enzyme Kinetics I – VI Introduction to Metabolic Control Analysis Flux Balance Analysis I & II NOTE: all other interests during course (introduce appropriately and constructively throughout) Mevalonate pathway Shikimic Acid Pathway Methyl-Erythritol Phosphate Pathway Phenylpropanoid Pathway Polyketides Prerequisites: Biochemistry I, Organic Chemistry I Organic Synthesis Laboratory Practice of organic laboratory techniques. Three hours of laboratory per lab session, twice a week. Approved chemical safety goggles meeting whatever national standards. The purpose of this laboratory course is to introduce students to the techniques that organic chemists (as well as biochemists, physical chemists, etc.) use in their daily routines. After learning and understanding those techniques, students will apply their knowledge to new situations to understand synthesis reactions, molecular structure determination, and analysis of (un)known compounds. Organic chemistry laboratory is important for several reasons. It introduces students to many different laboratory practices and concepts that will be used in subsequent chemistry laboratory classes and in other laboratory situations in biology, pharmacy, and chemical engineering (just to name a few!). It is anticipated that by the completion of this course, students will be familiar with all of the following topics and techniques: Safety in the laboratory Interpreting and following scientific directions Keeping a proper lab notebook Names and proper usage of lab instruments Understanding of general properties of compounds (including solubility, miscibility, acid/base chemistry, etc.) Proper usage of glassware Isolation and purification techniques (including filtration, solvent removal, drying solutions, distillations, chromatography (thin-layer, column, and gas) and crystallization/recrystallization) Characterization techniques including spectroscopy and melting point determination Interpretation of scientific results including percent yield and recovery, melting point, boiling point, IR and NMR spectra, and Rf values Required Materials: A laboratory notebook with carbon(less) pages Approved safety goggles Lab coats Lab manual will be posted through Blackboard Typical text: C.F. Wilcox, M.F. Wilcox, "Experimental Organic Chemistry, A Small-Scale Approach", (3rd edition, 2010). Apart from use of software in lectures, students will use software to accompany experiments that provide detailed molecular/compound structure, target sites, functional groups, etc. etc. Such exhibits will accompany lab reports. Lectures --> Lecture sessions are designed to clarify the concepts covered in the lab, as well as give an overview of techniques that will be used in the lab. Attendance is expected: The labs are only 3 hours in duration, so these lectures will be where you learn everything that you’ll need. Lab exercises will be available on Blackboard for each week. Please be considerate of your fellow students during the lecture period. Disruptions of any kind will not be tolerated and may result in expulsion from the classroom. Laboratory --> You will be required to have appropriate clothing before being allowed to enter the lab. Pre-labs are due at the beginning of the lab, and results and postlabs are due at the beginning of the lab 1 week after completion of the experiment! You will be expected to adhere to all of the lab safety rules. You are all expected to do your part to maintain a clean lab environment as part of GLP (Good Lab Practices): All reagent and solvent bottles should be completely closed immediately after use; All spills and dribbles should be cleaned immediately; All glassware should be put away at the end of the lab, and walkways should be kept free of debris. The following is the distribution of possible points in the course: Library Searching Exercise Database Search Exercises (Spectroscopy and Chromatography) Lab Quizzes Reaction/Synthesis methods knowledge Appropriate choice of method Appropriate constituents and tools. Procedure/steps (summary and/or ordering) Stoichiometry problems Spectroscopy and/or Chromatography analysis/interpretation Applications and industries Multistep Reaction/Synthesis Labs Lab Cleanliness Pre-lab Submissions Lab Notebook and Reports Lab Final Day 1: Much resemblance to quizzes Day 2-3: Augmented with the following: Molecular modelling software exercises Two or Three Practicum Group Labs (open notes) Part A. Points deducted for incompetent questionnaire for safety procedures for respective lab Part B. 2-3 labs to be implemented with competent data recording and lab reports. YOUR LAB REPORT CONSISTS OF THREE (3) PARTS --> Part I - Prelab Report. A copy of your lab notebook pages containing the lab write-up and answers to any prelab questions. This is due at the start of each experiment. Part II - Results. A copy of your notebook pages containing observations noted during the lab experiment. Is due with Part III one week from the conclusion of the experiment. Part III - Postlab Report. A summary of results and answers to postlab questions. This can be written on separate loose-leaf paper. Is due with Part II one week from the conclusion of the experiment Course Outline: Week1 Check-in/Safety Video/ Safety Procedures and Regulations Fractional Distillation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation, results and analysis Week 2 Measuring the Melting Points of Compounds and Mixtures Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 3 Purification by Recrystallization and Melting Point Measurement Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 4 Nucleophilic Substitution: Synthesis (SN1 Mechanism and SN2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 5 Oxidation of Alcohols (Primary, Secondary and Tertiary). Infrared Spectroscopy. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Infrared Spectroscopy Results and analysis Week 6 Elimination Reaction (E1 Mechanism and E2 Mechanism) Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 7 Synthesis of Aspirin. Chromatography and/or Spectroscopy Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Chromatography and/or Spectroscopy Results and analysis Week 8 Solvent Extraction Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 9 Electrophilic Aromatic Substitution: Synthesis of o- and p-Nitrophenol. No distillation; extract product with ethyl acetate. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 10 Separation and purification of o- and p-Nitrophenol by Liquid Chromatography. Use 100 mg sample, check by chromatography. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Results and analysis Week 11 Aldol Condensation Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 12 Grignard Reaction: Synthesis of Phenylmagnesium Bromide. Week 1: Part 1. Add methyl benzoate and sustain the desiccator for next week. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 13 HCl workup of previous week’s product. Synthesis of Triphenylmethanol and recrystallization of product. Purity check by melting point measurement. Concept Applications in industries Logistics and safety Molecular modelling simulation with software Lab implementation Consider other physical and chemical properties, such as solubility and spectroscopic data, to confirm the identity of the unknown compound. Results and analysis Week 14 -15 Wrapping up/cleaning things up. Final Exam. Prerequisite: Organic Chemistry I Functional Genomics This course will focus on execution of tools and protocols used to elucidate the biology, ecology and life histories of organisms through analysis of their genomes. Using genome research projects recently completed by the instructor and collaborators or other entities as templates, students will carry out each step of the research pipeline (unique to that project) in depth – from taxon selection, bioinformatic analysis of next-generation sequencing data, genome assembly, gene prediction/functional annotation, and finally how these data answer a functional biological hypothesis or question about the target organism. As new topics are introduced, they will be framed in the context of their applicability to the current project. At the end of the course, students will be able to select the appropriate protocols and analysis pipelines to complete the majority of today’s genome research. Each topic (or introduced research project) will consist of one or more lectures with background discussion, tool and material review, and a second hands-on computer (or dry) lab period during which students will apply the concepts and tools to complete a phase the genomic analysis. This course illustrates evolutionary concepts current tools in a hands-on manner within the confines of a goal or results-oriented genome research project. Course Goals --> Apply knowledge of genome sequencing and bioinformatics analyses to test hypotheses regarding organismal biology and evolution Communicate effectively how various bioinformatic analyses are impacted by similarities and differences in eukaryote genomes Apply appropriate bioinformatic tools and analysis protocols for individual genome sequencing projects Analyse and report on recent published research in the field of genomics Readings --> Selected scientific articles, book chapters, and writings from the popular press. Each lab will use a research paper authored by whosoever as a model for the analyses to be carried out with expected results Software --> R and packages of interest (BiocManager, ChemmineR, dPCP, MAPITR) Thodberg, M., & Sandelin, A. (2019). A step-by-step guide to analysing CAGE data using R/Bioconductor. F1000Research, 8, 886 Cao, Y., Charisi, A., Cheng, L. C., Jiang, T., & Girke, T. (2008). ChemmineR: a compound mining framework for R. Bioinformatics (Oxford, England), 24(15), 1733–1734 < manuals.bioinformatics.ucr.edu/home/R_BioCondManual > < cran.r-project.org/web/packages/BiocManager/vignettes/BiocManager.html Plant breeding and genomics < https://plant-breeding-genomics.extension.org/r-tutorials/ > NOTE: will like to incorporate the R environment into labs as compliment to analyses. Paper discussion --> Students will present a recent genome paper on a topic of their choosing and recap the methods used and conclusions inferred from the analyses. Project Part A --> Students will assemble, annotate and describe a mock genome project using public and/or simulated sequence data. Project Part B --> Apart from plants and trees serving as sustenance and nutrition for ecological consumers, various unique plants and trees are known to produce unique chemicals for defence, regulation, ecological advantage, etc, etc. Interest examples of concern Psychoactive Hallucination Paralysis Poison Blindness Blisters (mouth sores) Saps Burns (second and third degree) Anaesthetic Antiseptic Stinging nettle Tissue Digestion/Carnivorous (pineapple, insect trappers, etc., etc.) Involves taxonomy process of identification and classification, genome recognition, genetic markers related to the metabolic process for the syntheses of such chemical activity. Pathways and biochemical processes for mechanisms and responses. Includes the chemistry that takes place for the infamous or highly regarded activities, with the effects and symptoms; accompany with Organic Chemistry and Biochemcal software use for demonstration. Project Part C --> --In the wild there are various plants and trees that never garner any particular attention in horticulture and agriculture. Student groups with collect leave samples of at least 3 leaves of such plants, and at least 3 leaves of such trees. Will have lab DNA sequencing activities towards identification of genomes; identify possible using genetic markers to compare with databases. --Will gather samples of diseased specimens towards DNA sequencing to search for genetic variations and/or mutations that may play a role in the development or progression of a disease. The disease-causing change may be as small as the substitution, deletion, or addition of a single base pair or as large as a deletion of thousands of bases. Assessment --> Paper discussion: 10% Mid-term: 30% Final exam (cumulative): 30% Project (A + B + C): 30% Topics --> --Introduction / Why sequence genomes; next-gen sequencing --Basics I: Genome/transcriptome assembly; gene prediction and annotation --Basics II: Sequence alignment, phylogenetics, phylogenomics --Lab I: Illustrating the basics --The algal tree of life; origins of photosynthesis – The Cyanophora paradoxa genome --The MAT hypothesis; plastid establishment – The Cyanophora paradoxa genome --Lab II: Working with C. paradoxa genome data --Extremophilic vs. mesophilic genomes; horizontal gene transfer (HGT) – The Porphyridium purpureum genome --Lab III: Working with P. purpureum genome data --Genomic adaptation to environmental stress; HGT revisited – The Picochlorum spp. Genome --Differential expression: Lipid production, nitrogen starvation in Phaeodactylum tricornutum --Differential expression: Lipid production, nitrogen starvation in Phaeodactylum tricornutum --Lab VI: Differential expression analyses illustrated with P. tricornutum data. --Metabolism and physiology inferred from genome data – The Gambierdiscus caribaeus genome --Lab VII: Metabolic pathways, KEGG and gene ontologies illustrated with G. caribaeus data. --Paper discussion I --Paper discussion II --Single nucleotide polymorphisms and directed evolution – Chlamydomonas reinhardtii resequencing --Lab VIII: Calling SNPs and estimating functional consequences --Genetic divergence and natural selection as evidenced in genome data. --Phylogenetic networks and alignment-free methods to illustrate shared ancestry Prerequisites: General Botany, Plant Physiology, Biostatistics I & II, Plant Biochemistry Cell Biology: The standard definition of a cell in most introductory biology texts includes the line that cells are “the fundamental building blocks of all organisms.” Because of this fact, trite though it may be, a detailed understanding of the fundamental processes of cellular function is critical to all specialties within biology, clinical or academic. Some of these processes, including for example the biochemical mechanisms underlying cellular energetics, are remarkably consistent from bacteria to human. Other cellular processes and structures vary from cell type to cell type or organism to organism, allowing for unique adaptations of cells and organisms to particular functions. For example, nerve cells have various properties allowing them to conduct electrical signals and therefore process information, while kidney cells are specialized for the secretion of waste, and red blood cells for the transport of oxygen and carbon dioxide. What are the differences in physiology from cell type to cell type determining these specific functions? During the first half of the semester we will focus primarily on the biochemical processes that underlie cellular function, with an emphasis on protein structure and function, ion transport mechanisms and energy metabolism. The second half of the semester will emphasize more the function of particular organelles, including cell membranes, intracellular compartments and the cytoskeleton, and the relevance of these structures on processes like cell signaling and mitosis. Throughout the course, we will emphasize how variability in these processes imbues different cell types with their unique functional abilities. We will also seek to understand the experimental evidence for the different facts and concepts we study: How do we KNOW that nerve cell signaling, for example, involves the release of neurotransmitters? Some of this experimental evidence will be explored in a hands-on way in the lab sections, some will be discussed during lecture, and some will be the subject of analysis in the reading of original scientific manuscripts. Finally, we will examine how malfunctions in the cellular processes we are studying underlie certain diseases. In particular, the final few lectures of the course will focus on the biology of cancer cells: how do changes in cellular processes allow cancer cells to proliferate and metastasize? What are some of the current clinical approaches to curing cancer by blocking or reversing these processes? Aspirations --> To understand fundamental concepts of cellular function. To understand, and be able to critically analyze, the scientific evidence underlying our current understanding of cellular processes. To develop skills, through lab experiments, in some of the specific methodologies used in the study of modern cell biology. To become skilled at formulating and testing hypotheses using these methods. To develop a preliminary ability to read and analyze the primary scientific literature: What are the major findings of a science paper? What evidence is presented to support these findings? Are there shortcomings, either in the methods used or the logic of the experiments, which might lead one to question the conclusions reached by the authors? To be able to put this knowledge into larger contexts of how disease states occur or how organisms function adaptively within their environments. Typical text: The World of the Cell, by Becker, Kleinsmith, and Hardin, 6th edition (2006), Pearson/Benjamin Cummings Class Requirements and Grading --> 1. Class Participation (10%) To include attendance, responses to questions I pose in class, participation in discussions, and simply raising your hand from time to time to ask questions or make a comment (something I DO expect you to do). 2. Quizzes (10%) Two short in-class quizzes during the first half of the course. 3. Homework/problem sets (5%) There won’t be many of these; I’ll assign them when we hit subjects that are especially involved to help you learn the material and to make sure everyone is on track. 4. Primary literature readings (10%) We will read two papers from the primary scientific literature during the second half of the course. In both cases there will be an in-class discussion of the paper and a “reading guide” set of short essay questions which will be graded. The first reading guide assignment will be due AFTER the in-class discussion; the second assignment will be due BEFORE the in-class discussion. 5. Laboratory reports (25%) The specifics of each week’s lab report will be discussed during lab section. Typically, each week’s lab report will be due the following Monday in lecture. 6. Mid-term exam (20%) TENTATIVE format to include an in-class component, a short oral component, and a take-home component. 7. Final exam (20%) LABS --> Lab instructions for each week will be handed out ahead of time, either distributed as hard copies in lecture or posted on the course Angel site (or both). You are responsible for reading the instructions before lab. Otherwise, labs tend to run late, you will have difficulty obtaining the necessary data and knowing what to do with it. Do not expect the instructor to go over every step of the lab procedure before you start. Labs will make great emphasis on strong, practical and constructive immersion into the following software to accompany hands-on activity: << VCell, TiQuant + TiConstruct + TISIM >> Such software provides strong quantitative/computational microscopic assessment of specimens (or whatever) at professional standards. Such provides better means of objectives and expectations towards hands-on labs. Each lab will be associated with an explicit lab report assignment (contained in the lab instructions), usually due in lecture the Monday following lab. Usually, you may either submit the report with your lab partner or independently. If a report is submitted jointly, both partners must have contributed equally, as per Honor Code responsibilities. Do NOT make the mistake of dashing off reports the night before in a single draft. These reports will collectively account for 25% of your course grade, so take them seriously. The lab is a potentially dangerous place and you are required to follow all instructions given by your lab instructor and presented in the lab instructions. Disregarding instructions, or coming to class late or unprepared, may result in grade penalties, in addition to being just plain dangerous for yourself and those around you. Note: students can apply stationary video recording of labs with assigned regulations (to be given). Course Topics: Chapter 1 -19, 24. Some topics will require at least one week of instruction. Labs --> Cell Culturing, Aseptic Technique Cell Culture: basic techniques, population curve Cell Counting and splitting plates Cell Staining Histology Electron microscope Cell Harvesting & Cell Lysis Fractionalization of cells Common method(s) will be implemented Discussion and logistics for immunomagnetc separation & magnetic beads Isolation of erythrocyte membrane proteins Analysis of erythrocyte membrane proteins Bradford Assay (also identifying advantages and disadvantages) SDS-PAGE Chloroplasts and the Hill reaction Prerequisites: General Biology I & II Plant Molecular Biology The first objective of this course is to acquire a working knowledge of the molecular biology of plants (current times), being broadly photosynthetic organisms. Emphasis placed on genes/genomes and processes that are unique, or of particular importance to plants; yet to treat the “plant counterpart” to particular universally important processes, such as nuclear transcription . The second objective is to focus attention on what constitutes important ongoing research in plant molecular biology, and how does it compare to similar research on non-plant systems. Of consequence, you will learn about great successes with plant research as well as some of the current barriers to discovery and exploitation of plants. Typical course text --> Biochemistry and Molecular Biology of Plants, edited by Buchanan B., Gruissem W., and Jones R. American Society of Plant Physiologists. Lab Guides --> M.S, Punia. (2018). Plant Biochemistry and Molecular Biology: A Laboratory Manual. Scientific Publishers Carson, S. et al. (2019). Molecular Biology Techniques: A Classroom Laboratory Manual. Academic Press Schuler, M. A. and Zielinski, R. E. (2012). Methods in Plant Molecular Biology, Academic Press Maliga, P. (1995). Methods in Plant molecular biology: A Laboratory Course Manual.(A Cold spring Harbour Laboratory Course Manual). Cold spring Harbour Laboratory Press Stanton B. Gelvin (1989). Plant Molecular Biology Manual. Springer Netherlands Emphasis in Software Immersion and Skills Enforcement --> There are various software that will serve well to this course that further encourages a modern and profession environment, extending beyond memor based studies. Will make emphasis with practically and constructively implementing software alongside labs. Likely, one particular software will not have all the qualities on e is interest, however, out of the following sets choosing a max of 2-3 in usage will be constructive << VMD (with NAMS), GROMACS, VOTCA, Desmond, UCSF Chimera, Molsoft, CCPN, RedMD + RedMDStream >> << BLAST, Unipro UGENE, Bioclipse, Staden Package, Bioconductor >> << EMBOSS + JEMBOSS + Pise + wEMBOSS + EMBOSS-Explorer >> Assessment --> 4 Exams 60% Labs 40% Labs --> Labs will pursue Molecular Biology experiments for plantae augmented with software use. Other labs concern replication of chosen journal articles. Course Outline --> The Genetic Engineering of Plants: Transforming the Nucleus and Chloroplasts The Molecular Biology of Plant organelles Chloroplast Biology and Gene Structure (multiple sessions) Chloroplast Genome Evolution and Expression (multiple sessions) Organelle Self-Splicing Introns and Horizontal DNA transfer Chloroplast Gene Regulation – External & Nuclear Control Mitochondrial Genome Structure and Expression (multiple sessions) RNA Editing and Organelles Targeting of Proteins to Chloroplasts & Mitochondria The Nuclear Genome of Plants Size & Composition DNA Instability: Transposable Elements DNA Repair Gene Regulation in Development Gene Expression and Regulation PTGS (RNA Silencing & MicroRNAs) Photomorphogenisis (Leaf Development) Flower Development: Homeotic Genes The Molecular Basis of Stress Response Responses to Abiotic stress (multiple sessions) Heat & Cold Anaerobiosis, oxidative Responses to Biotic Stress (i.e., Pathogen attack) Prerequisites: Cell Biology, General Botany, Plant Physiology, Biochemistry, Organic Chemistry I Plant Pathology A study of the nature and causes of disease in plants, emphasizing the principal diseases in common plants and crops. There will be two hours of lecture and four hours of laboratory per week. Upon completion of the course the student will be able to: 1. Describe the concepts of what constitutes disease in plants. 2. Identify major principles of plant pathology. 3. Recognize the etiological agents of disease. 4. Employ methods to diagnose and manage a wide range of plant diseases. 5. Describe aspects of integrated pest management. 6. Explain the impact of plant disease on human affairs. 7. Ecological effects of pest management and resolutions NOTE: any field collection will be done in rain or sunshine Literature: Agrios G. 2005. Plant Pathology 5th edition Elsevier Academic Press. 922 pages Lab Tools: Will be given materials and instructin literature Note: all labs A through M will be done Applied Resource: FAO AGRIS Assessment 3-4 Quizzes 3 Exams Labs Topic course Outline -Introduction to Plant Pathology -Concepts of Disease -Stages in the Development of Disease (Disease Cycle) -How Pathogens Attack Plants -How Plants Defend Themselves -Fungi as Plant Pathogens -Eubacteria and Atypical Procaryotes as Plant Pathogens -Nematodes as Plant Pathogens -Viruses as Plant Pathogens -Mealy bug and similar pestilence -Parasitic Higher Plants as Plant Pathogens -Physiological or Abiotic Diseases -Control of Plant Diseases LABS --> NOTE: Some labs will also incorporate preparing of slides of plant tissue and pathogen specimen. Pathogens and diseases focus may be altered due to environment one resides in. A. Plant Disease Walk: Recognition of healthy and diseased plants is important. It is also essential to know the difference between a sign and a symptom The objective of this exercise is to find some plant diseases. The first step in this process is to familiarize yourself with diseased plants that may be in your area. Look online or in a plant disease book beforehand to find some possible plant diseases that may be in your area. Pictures can be saved for reference later. Materials needed: pen or pencil, paper, and a 10X hand lens or magnifying glass. (Hand lenses can be found at bookstores or online.) Procedure: Now go outside and begin walking around, whether it is in your backyard, a field, or a park. Look at the plants around you from the grass on the ground to the trees in the sky. Begin by looking at healthy plants. It is important to know what a healthy plant looks like to help determine when you have a sick one. Once you have found a healthy plant look for one that is out of the ordinary, such as one that is wilted or has yellowing leaves. Wilting can be associated with the plant’s inability to move water in the vascular system (xylem). Note the differences between the healthy and sick plants. Be sure to examine many different plant types. As you study these plants, list 5 symptoms you observed and 5 signs you examined. If you have a hand lens, observe closely at the plant specimen (maybe a leaf or stem). Might be able to see fruiting bodies and if you see fruiting bodies then you may have found a sign of the disease. Over the next few days (limited) note in your journal any changes with the diseased plants that you saw on your walk. Ask yourself some questions. “Have they changed in colour or shape?” “What do you think the cause of the sickness is?” “Is it related to weather, such as being too cold or too hot?” “Is only one plant sick or many plants?” Be sure to also take pictures throughout the exercise to help show what the plant looked like when you first saw it and what it looked like a few weeks later. B. Introduction to plant pathology and mycology part I C. Mycology part II D. Diseases caused by Ascomycota (4 labs) E. Diseases caused by Oomycota F. Fungal isolation techniques Part I and Nematodes G. Fungal isolation techniques Part II and abiotic plant damage H. Viral Diseases I. Bacteria part I J. Bacteria part II and Stem Rust Differential Set K. Parasitic seed plants L. Lab experiments with implementation of PCR, FISH, ELISA and IF: --Enzyme-Linked Immunosrbent Assay (ELISA) Note: choice can be different to specmen in articles Description of technique and process Clark, M. F. (1981). Immunosorbent Assay in Plant Pathology. Annual Reviews, Volume 19 pages 83 – 106 Copeland R. (1998) Assaying Levels of Plant Virus by ELISA. In: Foster G.D., Taylor S.C. (eds) Plant Virology Protocols. Methods in Molecular Biology™, vol 81. Humana Press Pataky JK, et al (2004). Ability of an ELISA-Based Seed Health Test to Detect Erwinia stewartii in Maize Seed Treated with Fungicides and Insecticides. Plant Disease. 88(6):633-640 Eibel, P., Wolf, G.A. & Koch, E. Development and evaluation of an enzyme-linked immunosorbent assay (ELISA) for the detection of loose smut of barley (Ustilago nuda). Eur J Plant Pathol 111, 113 (2005). Logistics Detecting pathogens for chosen plant speciment --Polymerase Chain Reaction (PCR) Schena, L., Duncan, J. M. and Cooke, J. E.L. (2008). Development and Application of a PCR-based ‘Molecular Tool Box’ for the Identification of Phytophthora Species Damaging Forests and Natural Ecosystems. Plant Pathology 57, 64–75 Aljawasim, B. and Vincelli, P. (2015). Evaluation of Polymerase Chain Reaction (PCR)-Based Methods for Rapid, Accurate Detection and Monitoring of Verticillium dahliae in Woody Hosts by Real-Time PCR. Plant Disease, Volume 99, Number 6 Lamarche J. et al. (2015) Molecular Detection of 10 of the Most Unwanted Alien Forest Pathogens in Canada Using Real-Time PCR. PLoS ONE 10(8): e0134265. Visnovsky, S. D. et al (2020). A PCR Diagnostic Assay for Rapid Detection of Plant Pathogenic Pseudomonads. Plant Pathology, Volume 69, Issue 7, pages 1311 – 1330 --Immunofluorescnce (IF) Wiwart, M., Mierzwa, Z. (1997). Indirect Immunofluorescence - an Useful Method in Studies on Some Fungal Pathogens. In: Dehne, HW., Adam, G., Diekmann, M., Frahm, J., Mauler-Machnik, A., van Halteren, P. (eds) Diagnosis and Identification of Plant Pathogens. Developments in Plant Pathology, vol 11. Springer, Dordrecht. Baysal-Gurel, F. et al. (2008). An Immunofluorescence Assay to Detect Urediniospores of Phakopsora Pachyrhizi. Plant Disease Vol. 92 No. 10 Janse J.D., Kokoskova B. (2009) Indirect Immunofluorescence Microscopy for the Detection and Identification of Plant Pathogenic Bacteria (In Particular for Ralstonia solanacearum). In: Burns R. (eds) Plant Pathology. Methods in Molecular Biology (Methods and Protocols), vol 508. Humana Press --Fluorescence In Situ Hybridization (FISH) Shakoori A. R. (2017). Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations, 343–367 Young, A. P., Jackson, D. J., & Wyeth, R. C. (2020). A Technical Review and Guide to RNA Fluorescence In Situ Hybridization. PeerJ, 8, e8806. M. Lab Final Exam Prerequisites: Cell Biology, General Botany, Plant Physiology, Field Botany, Biochemistry, Organic Chemistry I Forest Pathology The course consists of two lectures/labs (4 hours max) and, on average, 3-6 hours of field time per week. Lecture/lab time will be spent covering the biology, symptomatology and diagnosis of major fungal and bacterial diseases of particular trees, as well as mistletoes. Field identification and diagnosis is the focus of the course, with emphasis on late successional and old growth forests. Extensive required field trips will allow on-site assessment and identification of tree diseases, and quizzes and exams will take place in the field. Students are required to complete a collection of forest pathogens. Literature: Sinclair, W.A. & Lyon, H.H. 2007. Diseases of Trees and Shrubs. 2nd edition. Comstock Publishing, a Division of Cornell University Press, Ithaca, NY. 660 pp Additional: Tainter, F.H. & Baker, F.A. 1996. Principles of Forest Pathology. J. Wiley & Son, Inc. New York. 805 pp. Manion, P.D. 1991. Tree Disease Concepts. 2nd Ed. Prentice-Hall, Englewood Cliffs, New Jersey. 402 pp Assigned Readings --> Assigned readings will be made from the specified texts, research papers and Forest Service publications, etc. throughout the semester, and students are responsible for the material contained therein. Field Trips --> NOTE: will be done in rain or sunshine There will be 4-6 field trips Collection --> The purpose of the collection is to enable you to become adept at finding and identifying, on your own, forest pathogen signs, disease symptoms, and indications in the field. Rots (at least 3 on living trees) Rot diseases Rusts Foliage diseases Cankers/proliferations Mistletoes Labs --> Some labs will also incorporate preparing of slides of plant tissue and pathogen specimen. As well as identifying the biochemical/molecular biology process of such diseases/ailments. Incorporating lab experiments with PCR, FISH, ELISA and IF to be expected. Course Outline --> Introduction to forest pathology; the disease cycle Terminology: symptoms, signs, indications Wood Decay I. Brown Rots Wood Decay II. White Rots Root diseases I Root diseases II Rusts I Rusts II Foliage Diseases I Foliage Diseases II Cankers & Proliferations Mistletoes I Mistletoes II; completion of collections Prerequisites: Cell Biology, General Botany, Plant Physiology, Dendrology Forest Ecology Ecological interactions crucial to understanding forest ecosystems. Topics include: plant resources, competition, community development and dynamics, biodiversity, primary productivity, nutrient cycling, ecosystem structure and function, and impacts of global environmental change. Course examines numerous ways in which trees interact with their environments and influence ecological dynamics. We will investigate how trees sense and respond to environmental stimuli, shape patterns of biodiversity, influence ecosystem structure and function, and are impacted by global environmental change. As we focus on the science of forest ecology, we will also place strong emphasis on the professional development of each individual student. The course will offer significant opportunities to strengthen one's skills in thinking and communicating about scientific research and to consider how these skills can be employed in future careers. Each student will be expected to increase his or her understanding of best practices in scientific research and skills in formulating questions and synthesizing information. Course Literature When Forest Ecology is expressed I mean both plant and trees, with stimuli and other environmental agents. Treating only plants isn’t sensible. NOTE: course has a rain or shine policy with any outdoor activity Literature TBA Lecture Assessment: Quizzes Labs 3 Exams Projections and Species Distribution Project Projections and Species Distribution Project --> Will employ data sets for various ecosystems from unique regions; data sets subject to change. The given packages have vignettes to accompany their reference manuals; there may be supporting journal articles for each package as well. There must be competent overview and logistics to complement implementation for each packages. Students are required to apply the mentioned packages and given journal article. -simecol -popdemo -sdm or SSDM -Zhang L. et al. (2015) Consensus Forecasting of Species Distributions: The Effects of Niche Model Performance and Niche Properties. PLoS ONE 10(3): e0120056. Controversial topic research and presentation --> We can much more by having to teach and defend something to others, so you will have the opportunity to thoroughly research, present, and defend one of several topics relating to forest ecology to your peers, who will critically question both sides. Possible topics include: Transgenic Plants/Trees – Should plants be genetically modified for human use? Wildfire Suppression – Should wildfires be suppressed? Salvage Logging – Should salvage logging be allowed on federal lands? Logging Old-Growth Forests – Should logging occur in old-growth forests? Subsidization of Alternative Fuels – Should alternative fuels be federally subsidized? Forestry Based Carbon Offsets – Are forestry-based carbon offsets effective? Pre-European Settlement Benchmarks (PES) – Should PES be the benchmark for forest management and restoration efforts? Additional topics can be submitted by students until final date. On a given date all students MUST submit in order of preference three topics and whether they wish to work on the pro- or con-group. I will assign the topic, position and date to all students by whatever date. The pro- and con-position for each topic will be presented by a distinct group composed of 2-3 students. You may include economic, societal and political arguments into your discussion, but the primary focus MUST be scientific and if should directly relate to plant/forest ecology. One week prior to topic discussion a short paper (< 10 pages from either primary or secondary literature) supporting your group’s assigned viewpoint needs to be e-mailed to me or put in the appropriate “Dropbox” for the entire class to read. All papers turned in after the deadline will have 10 points deducted each day. I Course Outline: Individual Plant/Tree Interactions with Resources Light Water Mineral Nutrients & Multiple Limiting Resources Population Dynamics Population Structure & Plant/Trees Demography Species Life History Traits & "Strategies" Community Dynamics Community Assembly & Island Biogeography Succession Disturbance Species Interactions: Overview & Competition Species Interactions: Reproduction & Dispersal Species Interactions: Herbivory & Plant/Tree Pathogens Species Interactions: Plant/Tree-Soil Feedbacks Species Diversity Ecosystem Dynamics Decomposition & Soil Organic Matter Ecosystem Productivity Nutrient Cycling Human-Accelerated Environmental Change Deforestation Fire Ecology Invasions of Non-Indigenous Plants & Plant Pests Climate Change & Effect on Forests Overhunting & Overfishing Model developments based on data to exhibit cases Subject to real world constraints Invasive predators Invasive competitors Invasive consumers perturbating the ecological system Climate temperature acceleration with effects Predator-Prey Systems (ecological meaningfulness, NOT mathematical fetishes with asymptotics, chaos, bifurcations, Adjoint[Adjoint[Adjoint]], etc.). There will be much attempt to acquire real world meaningfulness for parameters w.r.t. to real data Lotka-Volterra Equations Generalised Lotka-Volterra Equations R packages of interest (deSolve, FME, primer) Restoration Ecology Controversial Topics Transgenic Plants/Trees Alternative Fuels Wildfire Suppression Salvage Logging Pre-European Settlement Benchmarks Climate Mitigation vs Adaptation Culling Class Party Labs --> Lab 1 Quantitative analysis Lab 2 Sapling growth responses to resources Field & Data work-up Lab 3 Forestry Forest succession: Field work Forest succession: Data work-up & writing Lab 4 – 5 Soil resources & forest community structure Field Trip to forest Effects of “whatever significant tree” on the establishment of inter-specific sdlg’s: Field Trip to Nature Preserve. “Whatever significant tree” lab: Field patterns, hypotheses, experimental design, writing successful research reports. Work on “whatever significant tree” experiments. Soil resources & forest community structure: Data work-up & writing. Lab 6 Perturbations Disturbance & species diversity: Field work. Disturbance & species diversity: Data work-up & writing Lab 7 Productivity Productivity: Data work-up & writing Lab 5 re-acquaintance. “Whatever significant tree” lab: Progress interview & Workshop (PowerPoint and presentation tips). Work on “whatever significant tree” experiments. “Whatever significant tree” symposium. Prerequisites: ODE, Biostatistics I & II, General Chemistry I & II, Field Botany, Dendrology Invasive Ecology Invasion Ecology is the study of introduced, non-native species and the factors that sometimes lead to their population explosions and negative ecological impacts in the new region. In this course, we will make explicit connections between fundamental concepts in ecology and evolutionary biology, topics specific to invasion ecology, and the idiosyncratic details surrounding particular invasive species. Goals for the course are to emphasize the ecological and economic importance of species invasions and to use the often-fascinating case studies from invasion biology to illustrate ecological and evolutionary principles. Layman terminologies that will resonate naturally throughout course: Pestilence Pathogens Parasitism Mass Consumption Disequilibrium Ecological feedback Trafficking Furthermore, concerning invasive ecology it’s not responsible to consider the botanic field concerns as disjoint from animalia in ecology. Ecosystems have equilibrium, dependencies, cooperation, etc., hence whatever invasive species present can perturb the ecosystem, influencing both producers and the levels of consumers, etc. By the end of this course, you should be able to: 1. Differentiate between commonly (mis)used terms used to describe introduced species. 2. Describe the major stages of, and barriers to, invasion success. 3. Describe major hypotheses used to explain invasion success. 4. Understand and explain fundamental concepts in ecology and evolutionary biology in the context of species invasions. 5. Critically assess claims regarding invasive species from the media. 6. Search for references to provide background material and support inferences, distinguishing between peer-reviewed and other sources. This course also has a service-learning component, for which all students will volunteer 8 hours assisting with invasive species management and/or monitoring and then reflect on their experiences. To facilitate this, a field day will be organised during the semester where we remove invasive species, set up long-term monitoring plots and collect baseline pre- and post-removal monitoring data. Literature: Elton, CE. 2000. The Ecology of Invasions by Animals and Plants. University of Chicago Press. Lockwood, JL, MF Hoopes and MP Marchetti. 2013. Invasion Ecology, 2nd edition (selected chapters). Wiley-Blackwell. Course Assignments --> Naturally, course assignments will be given throughout term. Will be lecture related. Exams --> This class will have both a midterm and a final exam. The midterm exam will cover topics from the first half of the class while the final will be comprehensive, covering information from throughout the semester. You will be tested on information from all aspects of the course (course readings, lectures and class discussions) and will be provided with a study guide and an in-class review to help you prepare. Exams may be multiple choice, true/false and/or short essay. Case Study Paper --> You will write a paper focused on the biology and ecology of a single non-native species (or group of closely related, ecologically similar species) of your choosing, based on peer-reviewed scientific literature. The objectives of this exercise will be to familiarize yourself with important characteristics of the species relating to its biology, the ecological role it plays in both the native and introduced regions, and the factors that facilitated its introduction to and establishment in the introduced region (referring to one or more of the major invasion hypotheses we’ve covered during the course). You will also be asked to identify key unanswered questions or areas of conflict in the literature regarding invasion success for which additional study would be useful. Case Study Poster Presentation --> Sharing research results with a larger audience is a critical component of the scientific process. You will prepare and present a poster and accompanying brief oral presentation to share your findings from the Case Study Paper with a general non-science audience in a poster session held at the end of the semester. You will be assessed based on your poster and oral explanation of your findings (40 points), as well as engagement with your fellow students about their work (10 points). Because the intended audience is the general public, this is an opportunity to be creative about how you share your findings. It will also allow you to compare-and-contrast your system with systems on which your peers have chosen to focus. Invaders --> in the News Introduced and/or invasive species are commonly mentioned in the news and popular press. Some well-known examples that you may be familiar with already. Over the course of the semester, you will identify three (3) examples of introduced species in the news and prepare a brief written summary of the story and why it is (or is not) important from an ecological and societal perspective (20 points each × 3 = 60 points). For one of these three stories, you will dig deeper into the specific claims made in the article and prepare a brief report that assesses the extent to which those claims are justified based on peer-reviewed scientific literature on the topic (40 points). The objective of this exercise is to think critically about how invasive species are portrayed in the popular press and assess how well those outlets tend to interpret scientific evidence for the general public. Computational & Empirical Development Projects (not necessarily in given order--> Peterson, A. (2003). Predicting the Geography of Species’ Invasions via Ecological Niche Modeling. The Quarterly Review of Biology, 78(4), 419-433 Robert F. Doren, Jennifer H. Richards, John C. Volin (2009). A Conceptual Ecological Model to Facilitate Understanding the Role of Invasive Species in Large-scale Ecosystem Restoration, Ecological Indicators, Volume 9, Issue 6, Supplement, Pages S150-S160 Crall, A., Jarnevich, C., Panke, B., Young, N., Renz, M., & Morisette, J. (2013). Using Habitat Suitability Models to target Invasive Plant species Surveys. Ecological Applications,23(1), 60-72. Lustig, A. (2017). Modelling Framework for the Establishment and Spread of Invasive Species in Heterogeneous Environments. Ecology and Evolution, volume 7 Issue 20, pages 8338 – 8348 Kariyawasam, C. S., Kumar, L., & Ratnayake, S. S. (2019). Invasive Plants Distribution Modeling: A Tool for Tropical Biodiversity Conservation with Special Reference to Sri Lanka. Tropical Conservation Science Roy-Dufresne, E., et al. (2019). Modelling the Distribution of aWwide-Ranging Invasive Species using the sampling Efforts of Expert and Citizen Scientists. Ecology and Evolution, 9(19), 11053–11063. Prerequisites: Biostatistics I & II, Forest Ecology Ecology Methods A survey of physical and biological processes that control the distribution and dynamics of vegetation and common methods used to understand these patterns. Through fieldwork and individual projects, students will gain hands-on experience regarding concepts and field methods in vegetation science. Based at (whatever) research station, we will spend 3 weeks exploring the ecology of the region – from tip to tip, with a focus on dynamics of various landscapes (mountainous, forest, plains, grasslands, etc, etc). Fieldwork will emphasize conceptual bases for and practicalities of vegetation research. NOTE: course will be given in “summer” and “winter” terms to not be in conflict with other courses. Course will run 3 - 5 weeks with “blue collar” hours each scheduled day. NOTE: students are expected to have proper apparel for environments to immerse into. Rehydration, eco-friendly sunscreen/bug repellent, head shade, insulated smartphone (not for music nor social media, nor passing time with entertainment), etc. etc. NOTE: It’s expected that students are competent with geographical data recording (latitude, longitude, elevation, date, time, province; such augment ecology data. Noteworthy: THIS IS A RAIN OR SHINE COURSE Course components – Background lectures Field trip (whatever altitudinal transect) Field exercises Readings Individual research projects Text: Mueller-Dombois & Ellenberg. 2003. Aims and Methods of Vegetation Ecology, Reprint of 1st edition, Blackburn Press. References: Texts from Field Botany course Texts from Dendrology course Tools: Google Maps Google Earth Assessment: Participation in field exercises (20%) Written assignments & in-class activities (45%) Project (design, implementation, and presentation) (30%) Field journal (5%) Specific objectives are to: Understand the goals and key concepts of vegetation/tree science Know how to design and implement a vegetation/trees field study Be familiar with general foundation information on common classification systems and ambiance vegetation/trees As a model for working in other ecosystems, be familiar with Colorado Front Range vegetation/trees in terms of (1) common species and plant lifeforms and (2) controls over landscape distribution of predominant vegetation types Identify common ambiance vascular plants General topics include: (1) Concepts of communities – complementary perspectives (2) Vegetation/tree classification systems – physiognomic, floristic approaches (3) Structure and function of ambiance biomes/ecoregions (4) Survey of factors controlling the distribution and dynamics of vegetation at continental, regional, landscape, and site scales – and ways to study vegetation at each of these scales Climate, physiography, soils, biotic interactions, time (succession, disturbance, etc.) (5) Concepts in field research design for Assessing vegetation/trees in space (classification, mapping, microhabitat studies) Assessment in time (vegetation dynamics, monitoring) Setting up hypotheses-driven experiments (6) Practicalities of field research Problem formulation Sampling protocol development (field technique selection, sampling design, etc.) Orienteering: map, compass, GPS, clinometer Supplemental data resources – vegetation-tree/soil maps, site histories Data management (QA) Data analysis (see Statistics) Communication of results (graphic visualization, oral and written presentation) (7) Plant/tree identification skills Major vascular plant/tree family characteristics Identification of common ambiance genera and species Use of dichotomous keys (8) Statistics – tools and design considerations Descriptive stats Exploratory stats – cluster analysis, ordination Hypotheses testing (9) How field studies interface with other areas in vegetation/landscape/ecosystem science Conservation, land management GIS, remote sensing applications Simulation (numerical) modelling – parameterization, validation Prerequisites: Field Botany, Dendrology, Biostatistics I & II, Upper Level Standing Geographic Information Systems for Biologists: The field of Geographic Information Systems, GIS, is concerned with the description, analysis, and management of geographic information. This course offers an introduction to methods of managing and processing geographic information. Emphasis will be placed on the nature of geographic information, data models and structures for geographic information, geographic data input, data manipulation and data storage, spatial analytic and modelling techniques, and error analysis. NOTE: course is focused mostly of botanical and ecological interests, with some climate/meteorological elements. The course is made of two components: lectures and labs. In the lectures, the conceptual elements of the above topics will be discussed. The labs are designed in such a way that students will gain first-hand experience in data input, data management, data analyses, and result presentation in a geographical information system. The basic objectives of this course for students are: 1. To understand the basic structures, concepts, and theories of GIS 2. To gain a hand-on experience with a variety of GIS operations Typical Texts: Longley P.A., M.F. Goodchild, D.J. Maguire, D.W. Rhind, 2011.Geographic Information Systems and Science. John Wiley and Sons Chang, K.T., 2012. Introduction to Geographic Information Systems (Sixth Edition). McGraw Hill, New York de Smith, M., Goodchild, M., Longley, P., 2013. Geospatial Analysis: A Comprehensive Guide (www.spatialanalysisonline.com) Tools --> A GIS of your choosing; students will be debriefed on operational requirements Google Earth Google Maps Resources --> https://www.google.com/earth/outreach/learn/ support.google.com/maps/answer/144349 There are highly established freeware GIS tools for use. Premier such available are SAGA GIS, ILWIS, MapWindow GIS, uDig, GRASS GIS and others; check Goody bag post. NOTE: GRASS GIS may be prerefence. Major priorities are sustainable skills in logistics, data management, accessibility & integration of data sets for project development and exhibition. Project(s) to have considerable life cycles with future use. Additionally, Google Earth and Google Maps can possibly coexist in such a instruction environment, primarily for rapid data visualisation. Course is concerned with the ability to develop meaningful professional data analysis and visualisation of sustainable value to whatever specified target audience. Unique talent development among such tools are encouraged, under the condition that the interests or demand of the target audience is appeased, of high quality. Some highly capable students will be able to develop projects with various systems, while for others finding an environment that suites them is key (highly dependent on what they comprehend and the effort they give). NOTE: in this course data from prerequisites can/will be applied to be constructive; dates and times must be instituted. Meteorological and oceanographic data may also be incorporated. Class Presentation --> Students need to review a journal article (or multiple articles) and give a presentation in the class. The article or articles can relate to GIS concepts, theories, or applications. An article in your discipline is preferred for you to review, for the reason that it would help you to think how to apply GIS in your work in the future. To present your reviewed article, you need to prepare five to eight slides in the format of PowerPoint, which would take approximately five to six minutes to present. In your slides, one of them would be how GIS is helpful in the article. You will have to give a small demonstration of some partial development for your project that substantially relates to your goals with whatever choice of tool employed. Followed by some substantial development (already done) with a GIS or other tool, or combination. You will have two or three minutes to answer the questions raised by the audience. Grading --> Lab Exercises 40% Exam I 20% Exam II 20% Presentation 20% Labs --> There are two components for labs: 1. Having GRASS GIS as preference concerns standard developments with course progression. 2. Extracurricular activities with Addons for GRASS GIS. Primarily, there must be strong development for a specific topic in (1) in order to commence with a respective Addons activity -- https://grass.osgeo.org/grass82/manuals/addons// Multicriteria decision decision analysis must be one topic for Addons extracurricular activities. An example: Massei, G., et al (2014). Decision Support Systems for Environmental Management: A Case Study on Wastewater from Agriculture, Journal of Environmental Management, Volume 146, Pages 491-504 However, PROMETHEE is not our only interest, and multiple MCDA addons will be pursued. Course Outline --> WEEK 1 Course Overview GIS Overview The Nature of Geographic Information WEEK 2 Data Representation Measuring Systems: Location – Coordinate Systems Data Representation Measuring Systems: Location – Coordinate Systems (Continue) WEEK 3 Data Representation Measuring Systems: Location – Coordinate Transformation Data Representation Measuring Systems: Topology Measuring Systems: Attributes WEEK 4 Data Representation Spatial Data Models: Introduction to spatial data models Spatial Data Models: Raster data models Data Representation Spatial Data Models: Relational Data Models Spatial Data Models: Vector Data Models (I) WEEK 5 Data Representation Spatial Data Models: Vector Data Models (II) Data Representation Spatial Data Models: TIN Summary of Spatial Data Models: Raster, Vector, TIN WEEK 6 Data Representation Linking attribute data with spatial data Recent Development of Data models WEEK 7 GIS Database Creation and Maintenance (I) Data Input & Editing GIS Database Creation and Maintenance (II) DBMS and its use in GIS WEEK 8 Review for Exam 1 Exam 1 WEEK 9 GIS Database Creation and Maintenance (III) Metadata / Database creation Guidelines / NSDI Data Analysis Measurement & Connectivity WEEK 10 Data Analysis Interpolation WEEK 11 Data Analysis Digital Terrain Analysis Data Analysis: Statistical Operations & Point Pattern Analysis WEEK 12 Data Analysis Classification Data Analysis GIS-based Modelling and Spatial Overlay (I) WEEK 13 Data Analysis GIS-based Modelling and Spatial Overlay (II) Data Analysis Summary Uncertainty WEEK 14 Geo-representation, Geo-presentation, and GeoVisualization GIS Applications WEEK 15 Student Presentations Student Presentations WEEK 16 Review for Exam Exam II Prerequisites: Field Botany, Dendrology, Aquatic Botany, Biostatistics I & II, Ecology Methods Plant Propagation I & II Principles and methods of plant propagation practiced. After completion of this class, you will have a practicable knowledge on seed germination and handling, rooting cuttings of various plant types, procedures for grafting and budding, using underground vegetative organs for plant increase, orchid propagation by rhizome and seed, and tissue culture propagation of selected plants. Students will acquire professional practice in plant propagation. Concerns being competent and professional with the implementation of grafting, cuttage, seedage and micropropagation. NOTE: this is a two term course. NOTE: Micropropagation will be treated extensively in the second term. The second term concerns interest in advance review and advance replication of labs of selected topics from prior term, but will be dominated by micropropagation pursuits. When successfully completing this course a student will be able to: Describe the principles of plant inheritance, plant reproduction and organogenesis. Recognise the physiological and anatomical changes a plant exhibits during asexual and sexual propagation. Select the appropriate methods of asexual and sexual propagation based upon biological characteristics of horticultural crops. Manipulate the propagation environment to promote the successful propagation of plants. Develop skills for using common asexual and sexual propagation techniques. Course Texts --> Plant Propagation: Principles and Practices, by Hartmann, H.T., D.E. Kester, F.T. Davies, and R.L. Geneve. Prentice Hall Plant Propagation; Concepts and Laboratory Exercises, 2nd Edition. Edited by Caula A. Beyl and Robert N. Trigiano, CRC Press Plants From Test Tubes, by Lydia Kyte & John Kleyn, Timber Press Course Assessment --> Quizzes Lab projects/experiments/reports 3 Exams Topics in the two term sequence --> Topics will be efficiently and tangibly administered to support lab activities given below. NOTE: for labs, if specimens are inaccessible or costly it’s possible to have more economic substitutes. Lab Activities -- Introduction to the greenhouse and propagation facilities, laboratory and safety procedures. Greenhouse procedures, and intermittent mist systems. Asexual Prop: Woody stem propagation Asexual Prop: Herbaceous stem propagation, leaf cuttings, root cuttings, & grass propagation Asexual Prop: Scaling of Lilium bulbs, twin-scaling of tunicate bulbs Sexual Prop: Orchid seed & fern spore germination Asexual Prop: Herbaceous plant grafting Asexual Prop: Woody plant grafting Asexual Prop: Geophytes Sexual Prop: Controlled pollination and hybridization. Sexual Prop: Seed dormancy and germination Propagation guides Micropropagation hands-on practical skills: Sterile technique Preparation of plant tissue culture nutrient media. Within vitro embryo culture (Flasking) and acclimatization techniques for growing orchids: Sterilization and sowing of orchid seeds. Learn the sequence of orchid seed germination and seedling development Planting out and managing orchid seedlings. Micropropagation via axillary shoot culture for propagation of Kalmia: Initiation Subculture and multiplication Ex vitro rooting and acclimation Lab Exercises in the given order: 1. Media Preparation 2. Initiation (Stage I) 3. Subculture and multiplication (Stage II) 4. Ex vitro rooting and acclimation (Stage IV) 5. Sterilization and sowing of orchid seeds 6. Subculture (reflasking) orchids 7. Planting out and managing orchid seedlings Prerequisites: at least upper junior Standing Wildlife Conservation Models Course introduces historical and contemporary advancements in the development of wildlife habitat models and their implementation in conservation planning. Course introduces current and effective techniques of wildlife modelling. Course will have a mixture of concepts, methodology, and applications topics towards development research and modelling for wildlife conservation. Course Literature --> Thompson, F. R. and Millspaugh, J. (2008). Models for Planning Wildlife Conservation in Large Landscapes. Netherlands: Elsevier Science Note: hopefully adjustment to ambiance of interest can be done. Tools --> GIS of choice (preferably GRASS GIS) Will be applying addons as well LANDIS II < https://www.landis-ii.org/home > Mentioned software in text will be introduced and implemented following analysis of their model structure and logistics for implementation; if software given are out of date or inaccessible, will then incorporate substitutes. USDA Natural Resources Conservation Service: Science and Technology - Conservation Tools Software (with analytical supporting documentation, then logistics overview before implementation) Wildlife Habitat Index RUSLE2 SPAW WEPS Win-PST NRL Bioenergy Models: https://bioenergymodels.nrel.gov/models/ Development in Course --> Course will be projects based where commitment and drive are crucial. Yet, there can be quizzes for concepts, developmental procedures and logistics. Labs --> Note: labs will likely require multiple sessions for completion. Groups may be assigned unique environments --Development with GRASS GIS mentioned in Tools. Due to prerequisites, development will be somewhat intensive. Will have heavy investment into Section II of course literature. --Habitat Networks for Terrestrial Wildlife --LANDIS II --USDA Natural Resources Conservation Service: Science and Technology - Conservation Tools Software (selection choices TBA) --GIS-Based Habitat Suitability Index (HSI) model (hopefully feasible in course, with need for data resources) --LCA in land use Chaplin-Kramer, R., Sim, S., Hamel, P. et al. (2017). Life Cycle Assessment Needs Predictive Spatial Modelling for Biodiversity and Ecosystem Services. Nat Commun 8, 15065 De Rosa, M. (2018). Land Use and Land-use Changes in Life Cycle Assessment: Green Modelling or Black Boxing? Ecological Economics, volume 144, pages 73 – 81 Prerequisites: Geographic Information Systems for Biologists SUMMER AND WINTER ACTIVITIES APPLICABLE TO BOTANY CONSTITUENTS: 3, 5, 15, 17, 18, 19, 20, 23, 24, 25, 26, 27, 29, 33, 37, 38, 39, 40, 42, 44 ADDITIONALLY --> The Forest Landscape Assessment Tool (FLAT) NOTE: students must successfully complete at least the Field Botany and Dendrology courses to participate in activity. Invasive Species and Pathology aspects likely to become inevitable with field observation (and among data collections), but such courses are not mandatory to participate in this activity. The Forest Landscape Assessment Tool (FLAT) is a set of procedures and tools used to rapidly determine forest ecological conditions and potential threats. FLAT enables planners and managers to understand baseline conditions, determine and prioritize restoration needs across a landscape system, and conduct ongoing monitoring to achieve land management goals. The rapid assessment process presents a cost-effective opportunity for landowners - including local governments, private owners, and nongovernmental organizations - to use ecological data to guide decision-making and improve environmental outcomes on their lands. Study Goals --> -Conduct forest assessment on over AB,000 acres of X County managed open space forest lands distributed across Y park sites -Establish baseline data that describes forest conditions per site and system wide -Identify key forest conditions that may need corrective and restorative actions -Develop long term forest stewardship recommendations for X County managers -Develop rapid forest assessment protocols that can be replicated on other public lands -Identify opportunities to collaborate with public & private agencies on forest stewardship Study Approach -Habitat Management Units (HMU) delineated for each site -Each HMU characterized by over story and under story species composition including invasive plants and other forest health indicators Ciecko, Lisa; Kimmett, David; Saunders, Jesse; Katz, Rachael; Wolf, Kathleen L.; Bazinet, Oliver; Richardson, Jeffrey; Brinkley, Weston; Blahna, Dale J. (2016). Forest Landscape Assessment Tool (FLAT): Rapid Assessment for Land Management. Gen. Tech. Rep. PNW-GTR-941. Portland, OR: U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station. < https://www.fs.fed.us/pnw/pubs/pnw_gtr941.pdf? > 1. Tools and Materials Before leaving on a field assessment A team should be properly equipped with required data collection tools. All field teams should have: Data entry tools Hand-held electronic data recorder or field data sheets Navigation devices GPS Map (including overlay of the MU boundaries) Compass Plant identification resources Camera Tree and canopy measurement tools For training and calibration of ocular estimate: Increment borer Diameter tape Densitometer (Moosehorn) Clinometer 2.Information Systems Software GIS (proprietary or open source, preferably GRASS GIS with addons) 3. THE “11” ESSENTIALS for comfort and safety in the field Sun protection (sunglasses, lip balm, and sunscreen) Bug repellent (eco-friendly) Proper clothing & footwear for harsh terrain or inclement weather such as rain gear, waterproof hiking/work boots, gaiters, and insulation like gloves, hats, and jackets. First aid supplies Utility knife or multi-tools (e.g. Leatherman, Swiss army knife) Food (plus an extra day’s supply) Lots of Water! (plus, an extra day’s supply) Headlamp or illumination source Fire (matches or lighter in waterproof container Emergency shelter (tent, tarp, bivy, or reflective blanket) Communication device smart phone or two way radio NOTE: don’t make activity a “Jurassic Park” event where one’s inconsiderate actions to have music jeopardizes themselves and others. NOTE: aerial imagery in terms of resource will be satellite imagery and data. Along with boundary, mapping and data, etc. NOTE: in the long run FLAT activities will reside in the same calendar often with FIA, REA and NRCA. FLAT Field Manual --> Parks and Recreation Division FLAT Field Manual: The Forestry Landscape Assessment Tool. Kings County Park, Department of Natural Resources and Parks https://forterra.org/wp-content/uploads/2015/06/FLAT_Field_Manual_Final-20131209.pdf Comparing plant cells PART A Students will observe plant cells using a light microscope. Two cells will be observed, one from the skin of an onion, and the other from a common aquarium water plant (anacharis or Elodea). Students will compare both types of cells and identify structures visible in each. Preliminary Elementary questions Name three structures found in plant/tree leaves cells AND in animal cells. Name two structures found in plant/tree leaves cells but not animal cells. What structure surrounds the cell membrane (in plants/tree leaves) and gives the cell support. What is the function of chloroplasts? Observation questions Describe the location and shape of chloroplasts. Why were no chloroplasts found in the onion cells? (hint: think about where you find onions) Determination of cell sizes Did you notice the chloroplasts moving within the cytoplasm of the elodea plant? Do they all move in the same pattern or direction? Suggest a reason why these structures move. Develop a quick experiment to test your hypothesis. Describe the test below, and if you have time conduct the test. PART B Pursue lab investigation based on the following: Wymer C.L., Beven A.F., Boudonck K., Lloyd C.W. (1999) Confocal Microscopy of Plant Cells. In: Paddock S.W. (eds) Confocal Microscopy Methods and Protocols. Methods in Molecular Biology, vol 122. Humana Press. PART D Metal shadowing and Freeze Fracture (if able) Visualize the surface of isolated subcellular structures or macromolecules in the transmission electron microscope. The specimen is coated with a thin layer of evaporated metal, such as platinum. The metal is sprayed onto the specimen from an angle so that surfaces of the specimen that face the source of evaporated metal molecules are coated more heavily than others. This differential coating creates a shadow effect, giving the specimen a three-dimensional appearance in electron micrographs. The preparation of samples by freeze fracture, in combination with metal shadowing, has been particularly important in studies of membrane structure. Specimens are frozen in liquid nitrogen (at -196°C) and then fractured with a knife blade. This process frequently splits the lipid bilayer, revealing the interior faces of a cell membrane. The specimen is then shadowed with platinum, and the biological material is dissolved with acid, producing a metal replica of the surface of the sample. Examination of such replicas in the electron microscope reveals many surface bumps, corresponding to proteins that span the lipid bilayer. A variation of free fracture called freeze etching allows visualization of the external surfaces of cell membranes in addition to their interior faces. Followed by biochemical and organic chemistry software to model and simulate macromolecules. Simulating the behaviour or reactions with common stimuli, penetration, etc, etc., etc. PART D Subcellular Fractionation Organelles extraction and centrifugation methods pursued towards observation PART E Plant cells can be grown and manipulated in culture. Such in vitro cell culture systems have enabled scientists to study cell growth and differentiation, as well as to perform genetic manipulations required to understand gene structure and function. Plant cells can also be cultured in nutrient media containing appropriate growth regulatory molecules. In contrast to the polypeptide growth factors that regulate the proliferation of most animal cells, the growth regulators of plant cells are small molecules that can pass through the plant cell wall. When provided with appropriate mixtures of these growth regulatory molecules, many types of plant cells proliferate in culture, producing a mass of undifferentiated cells called a callus. A striking feature of plant cells that contrasts sharply to the behaviour of animal cells is the phenomenon called totipotency. Differentiated animal cells, such as fibroblasts, cannot develop into other cell types, such as nerve cells. Many plant cells, however, are capable of forming any of the different cell types and tissues ultimately needed to regenerate an entire plant. Consequently, by appropriate manipulation of nutrients and growth regulatory molecules, undifferentiated plant cells in culture can be induced to form a variety of plant tissues, including roots, stems, and leaves. In many cases, even an entire plant can be regenerated from a single cultured cell. The ability to produce a new plant from a single cell that has been manipulated in culture makes it easy to introduce genetic alterations into plants, opening important possibilities for agricultural genetic engineering. PART F Xia Y., Petti C., Williams, M A. and DeBolt, S. (2014). Experimental Approaches to Study Plant Cell Walls During Plant-Microbe Interactions. Frontiers in Plant Science, Volume 5, Pages 540 Ecology Methods Reinforcement Reinforcement of field activities from course. There will be some preliminary Field Botany, Dendrology and Plant Systematics activity elements involved as well before implementing the ecology methods. Pesticide metabolism in Plants The following articles provide intelligence on pesticide metabolism in plants; however, interests also concerns experimental/laboratory pursuits. -Menn, J. (1978). Comparative Aspects of Pesticide Metabolism in Plants and Animals. Environmental Health Perspectives, 27, 113-124. -Van Eerd, Laura & Hoagland, Robert & Zablotowicz, Robert & Hall, J.. (2009). Pesticide Metabolism in Plants and Microorganisms: An Overview. Weed Science. 51. 472-495 -Katagi T. (2020). In vitro Metabolism of Pesticides and Industrial Chemicals in Fish. Journal of pesticide science, 45(1), 1–15. Xenobiotics metabolism in plants The following articles provide intelligence on xenobiotics metabolism in plants; however, interests also concern experimental/laboratory pursuits. Shimabukuro, R. H. and Walsh, W. C. (1979). Xenobiotic Metabolism in Plants: In Vitro Tissue, Organ, and Isolated Cell Techniques. Metabolism and Radiation Research Laboratory, Agricultural Research, Science and Education Administration, U. S. Department of Agriculture, Fargo, ND 58105 Sandermann H., Diesperger H., Scheel D. (1977) Metabolism of Xenobiotics by Plant Cell Cultures. In: Barz W., Reinhard E., Zenk M.H. (eds) Plant Tissue Culture and Its Bio-technological Application. Proceedings in Life Sciences. Springer, Berlin, Heidelberg. Sandermann H Jr. Plant Metabolism of xenobiotics. Trends Biochem Sci. 1992 Feb;17(2):82-4. Sandermann H. (1999) Plant Metabolism of Organic Xenobiotics. Status and Prospects of the ‘Green Liver’ Concept. In: Altman A., Ziv M., Izhar S. (eds) Plant Biotechnology and In Vitro Biology in the 21st Century. Current Plant Science and Biotechnology in Agriculture, vol 36. Springer, Dordrecht Rcompadre, Rage and popbio packages for botany and zoology Applying the R package Rcompadre and Rage to faciliate the use of the COMPADRE AND COMADRE databases and calculation of life history traits from matrix population models Owen R. Jones et al (2021). Rcompadre and Rage - Two R packages to Facilitate the Use of the COMPADRE and COMADRE Databases and Calculation of Life History Traits from Matrix Population Models. bioRxiv 2021.04.26.441330: https://www.biorxiv.org/content/10.1101/2021.04.26.441330v2.full.pdf Note: the Rcompadre and Rage packages are accompanied by vignettes; always observe the reference manuals as well because the vignettes don’t necessarily exhibit their full potential (which also involves your imagination and R skills) towards independent projects. Will pursue projects of interest. Note: the R package popbio can also serve well for additional interests. Observe its reference manual and the following: Stubben, C. and Milligan, B. (2007). Estimating and Analysing Demographic Models Using the popbio Package in R. Journal of Statistical Software, Volume 22, Issue 11 Such package also may have vignettes. Other R packages of interest (with analysis and logistics from supporting literature): -simecol -popdemo -sdm or SSDM Lab experiments with implementation of PCR, FISH, ELISA and IF Pathogens and diseases may be different to journal articles due to environment one resides in. Nevertheless, will also identify possible or ongoing threats in the ambiance. Will apply the mentioned techniques. Will also identify economic and ecologically friendly resolutions to such threats. --Enzyme-Linked Immunosrbent Assay (ELISA) Note: choice can be different to specmen in articles Description of technique and process Clark, M. F. (1981). Immunosorbent Assay in Plant Pathology. Annual Reviews, Volume 19 pages 83 – 106 Copeland R. (1998) Assaying Levels of Plant Virus by ELISA. In: Foster G.D., Taylor S.C. (eds) Plant Virology Protocols. Methods in Molecular Biology™, vol 81. Humana Press Pataky JK, et al (2004). Ability of an ELISA-Based Seed Health Test to Detect Erwinia stewartii in Maize Seed Treated with Fungicides and Insecticides. Plant Disease. 88(6):633-640 Eibel, P., Wolf, G.A. & Koch, E. Development and evaluation of an enzyme-linked immunosorbent assay (ELISA) for the detection of loose smut of barley (Ustilago nuda). Eur J Plant Pathol 111, 113 (2005). Logistics Detecting pathogens for chosen plant specimen --Polymerase Chain Reaction (PCR) Schena, L., Duncan, J. M. and Cooke, J. E.L. (2008). Development and Application of a PCR-based ‘Molecular Tool Box’ for the Identification of Phytophthora Species Damaging Forests and Natural Ecosystems. Plant Pathology 57, 64–75 Aljawasim, B. and Vincelli, P. (2015). Evaluation of Polymerase Chain Reaction (PCR)-Based Methods for Rapid, Accurate Detection and Monitoring of Verticillium dahliae in Woody Hosts by Real-Time PCR. Plant Disease, Volume 99, Number 6 Lamarche J. et al. (2015) Molecular Detection of 10 of the Most Unwanted Alien Forest Pathogens in Canada Using Real-Time PCR. PLoS ONE 10(8): e0134265. Visnovsky, S. D. et al (2020). A PCR Diagnostic Assay for Rapid Detection of Plant Pathogenic Pseudomonads. Plant Pathology, Volume 69, Issue 7, pages 1311 – 1330 --Immunofluorescnce (IF) Wiwart, M., Mierzwa, Z. (1997). Indirect Immunofluorescence - an Useful Method in Studies on Some Fungal Pathogens. In: Dehne, HW., Adam, G., Diekmann, M., Frahm, J., Mauler-Machnik, A., van Halteren, P. (eds) Diagnosis and Identification of Plant Pathogens. Developments in Plant Pathology, vol 11. Springer, Dordrecht. Baysal-Gurel, F. et al. (2008). An Immunofluorescence Assay to Detect Urediniospores of Phakopsora Pachyrhizi. Plant Disease Vol. 92 No. 10 Janse J.D., Kokoskova B. (2009) Indirect Immunofluorescence Microscopy for the Detection and Identification of Plant Pathogenic Bacteria (In Particular for Ralstonia solanacearum). In: Burns R. (eds) Plant Pathology. Methods in Molecular Biology (Methods and Protocols), vol 508. Humana Press --Fluorescence In Situ Hybridization (FISH) Shakoori A. R. (2017). Fluorescence In Situ Hybridization (FISH) and Its Applications. Chromosome Structure and Aberrations, 343–367 Young, A. P., Jackson, D. J., & Wyeth, R. C. (2020). A Technical Review and Guide to RNA Fluorescence In Situ Hybridization. PeerJ, 8, e8806. NOTE: other possible interests to develop Boothroyd, C. W., & Kelman, A. (1966). Laboratory Experiments in Plant Pathology. The American Biology Teacher, 28(6), 478–491. Structural Equation Modelling in Ecology Fan, Y., Chen, J., Shirkey, G. et al. (2016). Applications of Structural Equation Modeling (SEM) in Ecological Studies: an updated review. Ecol Process 5, 19 R environment assists: https://bookdown.org/bean_jerry/using_r_for_social_work_research/structural-equation-modeling.html https://quantdev.ssri.psu.edu/tutorials/structural-equation-modeling-r-using-lavaan https://stats.oarc.ucla.edu/r/seminars/rsem/ Ripeness and Staleness Also open to Biochemistry/Metabolic Biology students PART A (ripeness) Das, A. et al. (2016). Ultra-Portable, Wireless Smartphone Spectrometer for Rapid, Non-Destructive Testing of Fruit Ripeness. Sci Rep 6, 32504 Note: not interested in the additive manufacturing part, rather assembling primitive prototypes out of the components to implement. Note: generally use fruits attainable in your environment as substitutes. Side component 1: It’s important to characterise what type of molecules, polymers, etc. are distinctively prevalent for ripeness (for respective fruit). Considerable variation in freshness, say, groups consisting of consumable U were stored (G °C, I% RH) for 0, 2, 4, 6, 8, 10, etc., etc. days. Apply different types of consumables. Note: following such experiments to map the metabloc pathways for ripeneing. Can also include modelling and simulation of biochemical processes throughout. Side component 2: Experiments to conduct alongside side component 1 involving the same fruit speciments with 0, 2, 4, 6, 8, 10, etc., etc. days:-> Montero, T. M. et al (1996). Quality attributes of Strawberry During Ripening. Scientia Horticulturae, volume 65 Issue 4, pages 239 – 250 Ninio R. et al. (2003). Changes in Sugars, Acids, and Volatiles During Ripening of Koubo [Cereus peruvianus (L.) Miller] Fruits. J Agric Food Chem. 29; 51(3): pages 797-801 Anthon GE, LeStrange M, Barrett DM. (2011). Changes in pH, Acids, Sugars and Other Quality Parameters During Extended Vine Holding of Ripe Processing Tomatoes. J Sci Food Agric. 91(7): 1175-81. PART B (staleness) It’s important to characterise what type of molecules, polymers, etc. are distinctively prevalent for staleness (for respective consumable). Considerable variation in “age”, say, groups consisting of consumable X were stored (A °C, B% RH) for 0, 2, 4, 6, 8, 10, etc. etc. days. Apply different types of consumables. Note: generally use fruits attainable in your environment as substitutes. Note: there can be side component 2 like what is done in part A. Note: following such experiments to map the metabolic pathways for staleness. Can also include modelling and simulation of biochemical processes throughout. Wildlife Conservation Models Will be crash immersion OR advance recital of course detailed. Occupancy Modelling Note: will depend on data availability and credibility of such data. Guiding literature for activity --> MacKenzie, D. I. et al (2017). Occupancy Estimation and Modeling: Inferring Patterns and Dynamics of Species Occurrence. Academic Press Outhwaite, C. L., Chandler, R. E., Powney, G. D., & Collen, B., Gregory, R. S. & Isaac, N, J. B. (2018). Prior Specification in Bayesian Occupancy Modelling Improves Analysis of Species Occurrence Data. Ecological Indicators. 93. Pages 333-343 Stochastic Logistic Growth Models A. Review of deterministic standard logistic differential equation Model development and solution(s) Numerical Methods R packages B. Threshold Population Model Model development and solutions Numerical Methods R packages C. Extending (A) and (B) to Predator-Prey Models D. Stochastic logistic differential equation (SLE) Model development. Derivation (solution, expectation, variance) The distribution of SLE (with proof) Simulation by loop design. Investigate for different parameter values Use of R packages For simulation Monte carlo statistics at “final time T”. Mean path and confidence bands for this SDE Maximum and minimum values within confidence bands along with mean path. Analyse and develop: Møller, J. K., Madsen, H. and Carstensen, J. (2011). Parameter Estimation in a Simple Stochastic Differential Equation for Phytoplankton Modelling. Ecological Modelling 222, 1793 – 1799 Heydari, J., Lawless, C., Lydall, D. A., & Wilkinson, D. J. (2014). Fast Bayesian Parameter Estimation for Stochastic Logistic Growth Models. Bio Systems, 122, 55–72. NOTE: there may be other methods from other literature NOTE: will have other ecological interests E. Extending (D) to predator-prey models for ambiances of interest Phylogenetic Comparative Methods in R Revell, L. J. (2011). phytools: an R Package for Phylogenetic Comparative Biology (and other things). Methods in Ecology and Evolution 3(2), pp 217-223 Revell, L. J. and Harmon, L. J. (2022). Phylogenetic Comparative Methods in R, Princeton University Press Note: will pursue organisms of interest (botanic and animalia). US Government and IGO Resources tools for Agriculture MUST have analysis of supporting documentation before software use; followed by logistics, then software/tools implementation. Chosen software/tools sets subject to change. EPA WATERSHEDSS software: https://www.epa.gov/ceam/watershedss US FDA (1998) - Guidance for Industry: Guide to Minimize Microbial Food- Safety Hazards for Fresh Fruits and Vegetables USDA Integrated Farm System Model (IFSM) USDA Science and Technology Conservation Tools Software: https://www.nrcs.usda.gov/wps/portal/nrcs/detailfull/national/ndcsmc/?cid=stelprdb1042198 Other USDA software (huge lists): < https://data.nal.usda.gov/nal-terms/computer-software < https://www.ars.usda.gov/research/software/ Wauchope, R. D. et al (2003). Software for Pest-Management Science: Computer Models and Databases from the United States Department of Agriculture— Agricultural Research Service. Pest Manag Sci 59:691–698 OECD (1999), Environmental Indicators for Agriculture: Vol. 1: Concepts and Framework Vol. 2: Issues and Design -- "The York Workshop", OECD Publishing, Paris OECD (2001), Environmental Indicators for Agriculture: Vol. 3: Methods and Results. OECD Publishing, Paris 16. FSMA Final Rule on Produce Safety (country counterpart to): https://www.fda.gov/food/food-safety-modernization-act-fsma/fsma-final-rule-produce-safety NOTE: concerning field activities and labs for microbiology and botany the following text may serve well in backdrop: Millard, S. P. (2013). EnvStats: An R Package for Environmental Statistics, Springer
0 notes
Text
G2D2, 2: first week

The campus of the China Three Gorges Hotel has a small river, full of waterlilies and lotus flowers. Beside it runs a main road, along which big water trucks run, shooting atomised water from cannon-like structures; we are told this is to reduce the air pollution.
After an introductory party on Monday, the conference opened at 9:00 on Tuesday morning. In the first week, much of the activity is minicourses 1 and 2, the first on “Laplacian eigenvalues and optimality” given by Rosemary and me, the second on “Topics in representation theory” by Tullio Ceccherini-Silberstein.
But first, some words on the two logos, which appear at the top of the first post in this sequence.
The meeting is being held in the Three Gorges Mathematics Research Centre, whose logo is on the right. It is part of the China Three Gorges University, near the famous Three Gorges on the Yangtse River. The University’s logo, which I am sure your search engine will quickly find, features three vertical wavy shapes, which presumably are intended to suggest water cascading through the three gorges. The TGMRC logo cleverly replaces these random shapes by integral signs, which they somewhat resemble.
The TGMRC is on a corridor on the top floor of a U-shaped building called L1 or L2, depending on which side you enter. The top of the U is closed by a high bridge. So it somewhat resembles the new location of the ICMS in Edinburgh, except that that building is entirely enclosed, whereas the central courtyard of L1/L2 is open to sun and rain. There is a nice lecture room with comfortable chairs. Although there is no public space except for the corridor (where the welcome party was held), there are a number of rooms which can be used as “breakout rooms” for small discussions.
The conference logo on the left is a Deza graph. There has been a recent upsurge of activity on Deza graphs, which I suspect is driven by Sergey Goryainov.
A Deza graph is a regular connected graph for which the number of common neighbours of two vertices takes just two distinct values. If adjacent vertices have one number of common neighbours and non-adjacent vertices the other, then the graph is strongly regular. So a strictly Deza graph is defined to be a Deza graph of diameter 2 which is not strongly regular. The graph in the G2D2 logo is the smallest strictly Deza graph, as you can easily verify. I do not know whether the colours have any mathematical significance.
On Wednesday, Dmitry Panasenko told us about a clever computer search which found all the strictly Deza graphs on up to 21 vertices. For one parameter set (I think on 20 vertices) there are many non-isomorphic graphs (I think around 20 or 30, though I didn’t copy down the number from a long table). Then Soesoe Zaw told us about some new constructions of strictly Deza graphs from the Berlekamp–van Lint–Seidel graph, using the operation of dual Seidel switching due to Willem Haemers. This pleases me, since I am typing this on a computer called Seidel.
The Berlekamp–van Lint–Seidel graph is a graph on 243 vertices which is dual (in the sense of Delsarte duality for association schemes) to the graph associated with the perfect ternary Golay code C of dimension 5 over the field of 3 elements. Take the dual code of C (which is 6-dimensional of length 11 and contains C as a subcode of codimension 1). The vertices of the graph are the cosets of this dual code; two vertices are joined if their difference contains a word of weight 1. It is a “sporadic” strongly regular graph with 243 vertices, and from it Soesoe was able to construct a strictly Deza graph on 243 vertices and two of them on 486 vertices.
Seidel’s name came up again in Wei-Hsuan Yu’s talk on equiangular lines, in which he began with the results which I saw from close-up in the early 1970s by Lemmens, Seidel, Higman, Taylor, Gerzon, and so on, and then gave some improvements, some of which have been obtained by semi-definite programming; and again in Alexander Gavrilyuk’s talk on digraphs whose Hermitian adjacency matrices have spectral radius at most 2.
One of the invited speakers was Gareth Jones. He was going to tell us about his recent theorem that many triangle groups contain uncountably many maximal nonparabolic subgroups (subgroups maximal with respect to containing no parabolic elements in their action on the hyperbolic plane). But for reasons not revealed, he was unable to come to the meeting. Instead, Alexander Mednykh had offered to give a talk to Gareth’s slides. Very brave, I thought. But Sasha began by explaining to us the basic idea behind the proof, a mix of maps and hypermaps, coverings, Riemann surfaces, etc.
Qing Xiang gave us new bounds for the size of a partial ovoid in either the generalised quadrangle E(5,q) (what I might call Ω−(6,q) and in the Ree–Tits generalised octagon; in each case, previous bounds were of order a power of q, but he and his co-authors have managed to lower the exponent of the power. He showed us Chris Godsil’s nice proof of Jef Thas’ q3−q2+q bound in the GQ case, based on association scheme techniques (and so potentially useable in much more general situations). The new bound uses modulo p techniques and very specific results about dimensions of modules for algebraic groups, and while much more powerful in this case is likely to be much more specialised.
There have been several mentions of Cayley graphs, and I must point out a notational trap. Given a subset S of a group G, not containing the identity, the Cayley (di)graph Cay(G,S) is the graph with vertex set G and with an arc from x to sx for every s in S. If S is inverse-closed, it is an undirected graph. It admits the action of G on itself by right multiplication (the right regular action) as automorphisms. In fact, any graph which admits G acting regularly as a group of automorphisms is necessarily a Cayley graph for G.
Now there are two, quite different, definitions of a normal Cayley graph:
The definition I learned first says that Cay(G,S) is a normal Cayley graph if S is a normal subset of G (closed under conjugation by elements of G); equivalently, the graph admits both the left and the right regular action of G as automorphisms.
The definition which seems to be standard among participants at this conference is: Cay(G,S) is a normal Cayley graph if G is a normal subgroup of the full automorphism group of the graph.
The first definition is very restrictive but also very useful. For two examples,
A graph invariant under the primitive group of simple diagonal type whose socle is the product of two copies of the simple group T is a normal Cayley graph for T.
For n > 3, Henson’s universal homogeneous Kn-free graph is not a normal Cayley graph for any group (though Greg Cherlin showed that it is a Cayley graph).
The second definition, by contrast, says essentially that a normal Cayley graph has no “unexpected” automorphisms: its automorphism group is contained in the holomorph of G. In accordance with the principle that graphs tend to have few automorphisms other than those you must have, we would expect that most Cayley graphs are normal; this has indeed been conjectured, with some evidence.
I wish that we had different words for these two obviously useful concepts. But it is probably too late for that now; one of them will just fade away.
The paragraph two above reminds me of an old theorem of mine, the citation for which was just imported from Scopus into the St Andrews research repository. For every finite group G, there is a constant α(G), between 0 and 1, so that, in the class of n-vertex graphs whose automorphism group contains a subgroup isomorphic to G, the proportion whose automorphism group is exactly G tends to the limit α(G) as n→∞. In accordance with the above principle, you might expect that most groups have α(G) = 1; but in fact this happens if and only if G is a direct product of symmetric groups, and the values of α are dense in the unit interval.
The idea of terminology fading away came up in Misha Muzychuk’s nice survey of coherent configurations for the summer school. As I have probably pointed out here before, at the end of the 1960s, within a couple of years of each other, Donald Higman invented coherent configurations, while Weisfeiler and Leman invented cellular algebras. The latter notation has faded away, partly because the term has been re-used with an entirely different meaning. But long before this, Bose and Nair invented association schemes, which are an important special case of coherent configurations. There are four levels of generality; in increasing generality, a coherent configuration may be symmetric, commutative, homogeneous, or none of the above. Bose and Nair used the term “association scheme” for symmetric c.c.; Delsarte used it for commutative c.c.; Bannai and Ito used it for homogeneous c.c., and this seems to have become standard. I prefer to use the term “coherent configuration” with appropriate qualification.
Unfortunately Misha did not have time to describe his recent construction, with Klin and Reichard, of proper Jordan schemes; but Mike Kagan will talk later in the meeting, and I think he might say something about this.
from Peter Cameron's Blog https://ift.tt/2KHbSWq from Blogger https://ift.tt/2YVjXzn
0 notes
Text
I wish I was your derivative so I could lie tangent to your curves.
My love for you is like a concave up function because it is always increasing.
How can I know so many hundreds of digits of pi and not the 7 digits of your phone number?
I wish I was your second derivative so I could investigate your concavities.
You and I would add up better than a Riemann sum.
Hey baby, what’s your sine?
I need a little help with my Calculus, can you integrate my natural log?
By looking at you I can tell you’re 36-25-36, which by the way are all perfect squares.
You fascinate me more than the Fundamental Theorem of Calculus.
Are you a 90 degree angle? ‘Cause you are looking right!
My love for you is like pi… never ending.
I’d like to plug my solution into your equation.
Since distance equals velocity times time, let’s let velocity and time approach infinity, because I want to go all the way with you.
I am equivalent to the Empty Set when you are not with me.
I don’t like my current girlfriend. Mind if I do a you-substitution?
I can figure out the square root of any number in less than 10 seconds. What? You don’t believe me? Well, then, let’s try it with your phone number.
Hey, baby want to Squeeze my Theorem while I poly your nomial?
Hey…nice asymptote.
I’m not being obtuse, but you’re acute girl.
I don’t know if you’re in my range, but I’d sure like to take you back to my domain.
Are you a 45 degree angle? Because you’re acute-y.
My love for you is like y=2^x… exponentially growing.
I’ll take you to your limit if you show me your end behavior.
Can I explore your mean value?
The derivative of my love for you is 0, because my love for you is constant.
I’m good at math… let’s add a bed, subtract our clothes, divide your legs, and multiply!
Our love is like dividing by zero… you cannot define it.
If you were a graphics calculator, I’d look at your curves all day long!
I’ve been secant you for a long time.
If I’m sine and you’re cosine, wanna make like a tangent?
Meeting you is like making a switch to polar coordinates: complex and imaginary things are given a magnitude and a direction.
Being without you is like being a metric space in which exists a cauchy sequence that does not converge.
My love for you is a monotonically increasing unbounded function
Omg these were great!!!
2 notes
·
View notes
Text
Mathematics Plus Engineering Aptitude (Syllabus)
SYLLABUS PART A General skills with emphasis on reasonable reasoning, graphical analysis, synthetic and numerical ability, quantitative comparisons, series formation, questions, and so forth SYLLABUS PART B Mathematics Plus Engineering Aptitude Linear Algebra Calculus Complex factors Vector Calculus Ordinary Gear Algebra of matrices, inverse, rank, system of geradlinig equations, symmetric, skew-symmetric plus orthogonal matrices. Hermitian, skew-Hermitian and unitary matrices. eigenvalues and eigenvectors, diagonalisation associated with matrices. Functions of the solitary variable, limit, continuity plus differentiability, Mean worth theorems, Indeterminate forms plus L'Hospital rule, Maxima plus minima, Taylor's series, Newton’s method for finding origins of polynomials. Fundamental plus means value-theorems of essential calculus. Numerical integration simply by trapezoidal and Simpson’s guideline. Evaluation of definite plus improper integrals, Beta and Gamma functions, Features of two variables, limit, continuity, partial derivatives, Euler's theorem for homogeneous functions, total derivatives, maxima and minima, Lagrange technique of multipliers, double integrals and their applications, series and series, tests with regard to convergence, power series, Fourier Series, Half range sine and cosine series. Inductive functions, Cauchy-Riemann equations, Collection integral, Cauchy's integral theorem and integral formula Taylor’s and Laurent' series, Remains theorem as well as applications. Lean, divergence and curl, vector identities, directional derivatives, collection, surface and volume integrals, Stokes, Gauss and Green's theorems and their programs. First order equation (linear and non-linear ), 2nd order linear differential equations with variable coefficients, Variance of equation parameters technique, higher order linear gear equations with constant coefficients, Cauchy-Euler's equations, power collection solutions, Legendre polynomials plus Bessel's functions from the particular first kind and their own properties. Numerical solutions associated with first order ordinary gear equations by Euler’s plus Runge -Kutta methods. Meanings of probability and easy theorems, conditional probability, Bayes Theorem. Solid Body Movement and Fluid Motion: Energetics: Electron Transport: Electromagnetics: Materials: Particle dynamics; Projectiles; Rigid Body Dynamics; Lagrangian formulation; Eulerian formulation; Bernoulli’s Equation; Continuity equation; Surface area tension; Viscosity; Brownian Movement. Laws of Thermodynamics; Concept of Free energy; Enthalpy, and Entropy; Equation associated with State; Thermodynamics relations. The framework of atoms, Concept associated with energy level, Bond Concept; Definition of conduction, Semiconductor and Insulators; Diode; Fifty percent wave & Full influx rectification; Amplifiers & Oscillators; Truth Table. Theory associated with Electric and Magnetic possible & field; Biot and Savart’s Law; Theory associated with Dipole; Theory of Vacillation of electron; Maxwell’s equations; Transmission theory; Amplitude plus Frequency Modulation. Periodic desk; Properties of elements; Outcome of materials; Metals plus nonmetals (Inorganic materials), Primary understanding of monomeric plus polymeric compounds; Organometallic substances; Crystal structure and proportion, Structure-property correlation-metals, ceramics, plus polymers.
0 notes
Text
Returning to CSE maths 4 years after High School
I know this subreddit says that questions about "learning maths" should be on r/learnmath but I feel that my question is a little more focused (not just about...wanting book or resources) and could be answered here. Nevertheless, I will be posting this on there too.
This is a long one... bear with me. If you will.
I am going into the second year of a Computer Science degree and we have a course called "Engineering Mathematics" (ME3) in the next semester.
I graduated high school a WHILE ago and honestly need a little brushing up before I start learning ME3. But I don't have the time to go through all the maths topics we had then in all the 4 years. I was wondering if someone could help me decide what I should revisit and revise before going on to ME3.
Course Content of ME3
------------------------------------
1 - Linear Differential Equations (LDE)
\- LDE of nth order with constant coefficients \- Method of variation of parameters \- Cauchy's & Legendre's LDE \- Simultaneous & Symmetric Simultaneous DE \- Modelling of Electric Circuits
2 - Transforms
\- Fouriers Transform \- Complex exponential form Fourier series \- Fourier Integral Theorem \- Fourier Sine & Cosine Integrals \- Fourier Sine & Cosine transforms & their inverses \- Z Transform (ZT) \- Standard Properties \- ZT of standard sequences & their inverse
3 - Statistics
\- Measures of Central tendency \- Standard deviation, \- Coefficient of variation, \- Moments, Skewness and Kurtosis \- Curve fitting: fitting of straight line \- Parabola and Related curves \- Correlation and Regression \- Reliability of Regression Estimates.
4 - Probability and Probability Distributions
\- Probability, Theorems on Probability \- Bayes Theorem, \- Random variables, \- Mathematical Expectation \- Probability density function \- Probability distributions: Binomial, Poisson, Normal and Hypergometric \- Test of Hypothesis: Chi-Square test, t-distribution
5 - Vector Calculus
\- Vector differentiation \- Gradient, Divergence and Curl \- Directional derivative \- Solenoid and Irrigational fields \- Vector identities. Line, Surface and Volume integrals \- Green‘s Lemma, Gauss‘s Divergence theorem and Stoke‘s theorem
6 - Complex Variables
\- Functions of Complex variables \- Analytic functions \- Cauchy-Riemann equations \- Conformal mapping \- Bilinear transformation \- Cauchy‘s integral theorem & Cauchy‘s integral formula, \- Laurent‘s series, and Residue theorem
-------------------------------------------------------------------------------------------
Overview of roughly what all we had in the years of High School, a little out of order because I am summarizing all 4 years of Math books.
Real Numbers
\- Laws of Exponents for Real Numbers \- Euclid’s Division Lemma \- Fundamental Theorem of Arithmetic
Polynomials
\- Polynomials in One Variable \- Zeroes of a Polynomia, Remaider Theorem, Factorization of Polynomials \- Relationship between Zeroes and Coefficients of a Polynomial \- Division Algorithm for Polynomials
Pair of Linear Equations in Two variables
\- Linear Equations \- Solution of a Linear Equation \- Pair of Linear Equations in Two Variables \- Graphical Method of Solution of a Pair of Linear Equations \- Substitution Method, Elimination Method & Cross-Multiplication Method
Principles of Mathematical Induction
Complex Numbers
\- Modulus and the Conjugate \- Argand Plane and Polar Representation
Quadratic Equations
\- Factorisation & Completing the Square, Roots of Equations.
Sets----------
\- Sets: Empty, Finite, Infinite, Equal, Subsets, Power Set, Universal Set. \- Venn Diagrams \- Union, Intersection & Complement of a Set
Permutations and Combinations
Binomial Theorem
\- Binomial Theorem for Positive Integral Indices \- General and Middle Terms
Sequences and Series
\- Sequences & Series \- Arithmetic Progressions \- nth Term of an AP, Sum of n terms of an AP \- Geometric Progression \- Relationship Between Arithematic Mean and Geometric Mean
Matrices
\- Types & Operations \- Transpose \- Symmetric and Skew Symmetric Matrices \- Transformation \- Invertible Matrices
Determinants
\- Properties of Determinants \- Area of a Triangle \- Minors and Cofactors \- Adjoint and Inverse of a Matrix \- Applications of Determinants and Matrices
Relations and Functions
\- Cartesian Product of Sets \- Relations & Functions \- Composition of Functions and Invertible Function \- Binary Operations
Limits and Derivatives
\- Limits, Derivatives \- Limits of Trigonometric Functions \- Applications: Rate of Change of Quantities, Increasing and Decreasing Functions
Tangents and Normals, Approximations & Maxima and Minima
Continuity and Differentiability
\- Exponential and Logarithmic Functions \- Logarithmic Differentiation \- Derivatives of Functions in Parametric Forms \- Second Order Derivative \- Mean Value Theorem
Integrals
\- Inverse Process of Differentiation \- Methods of Integration \- Integration by Partial Fractions & by Parts \- Definite Integral \- Fundamental Theorem of Calculus \- Definite Integrals by Substitution \- Properties of Definite Integrals \- Applications: Area under Simple Curves, Area between Two Curves
Differential Equations
\- Basic Concepts \- General and Particular Solutions of Differential Equation \- Differential Equation whose General Solution is given \- Methods of Solving First order, First Degree Differential Equations
Vector Algebra
\- Types of Vectors \- Addition of Vectors, Multiplication of a Vector by a Scalar \- Product of Two Vectors
Linear Programming
Statistics
\- Graphical Representation \- Distribution. Mean, Mode & Median \- Measures of Dispersion, Range, Mean Deviation \- Variance and Standard Deviation
Probability
\- Random Experiments, Events, Axiomatic Approach to Probability \- Conditional Probability , Multiplication Theorem, Independent Events \- Bayes' Theorem \- Random Variables and their Probability Distributions \- Bernoulli Trials and Binomial Distribution
------------------------------------------------------------------------------
Euclids's Geometry
Properties of Lines, Angles, Circles, Triangles, Quadrilaterals, Parallelograms (Too easy to worry about)
Some chapters about Areas & Volumes of Quadrilaterals, Circles, Cylinders, Cuboids & Spheres (Again.. too easy)
Heron's Formula
\- Area of a Triangle – by Heron’s Formula \- Application of Heron’s Formula
Trigonometry
\- Trigonometric Ratios, Identities \- Applications : Heights and Distances
Trigonometric Functions
\- Sum and Difference of Two Angles \- Trigonometric Equations \- Inverse Trigonometric Functions & their Properties
Circles
\- Tangent to a Circle
Straight Lines
\- Slope of a Line \- Forms of Equations of a Line \- Distance of a Point From a Line
Conic Sections
\- Cone, Circle, \- Equations: Parabola, Ellipse & Hyperbola \- Eccentricity, Latus rectum
Three Dimensional Geometry
\- Coordinate Axes and Coordinate Planes in 3D Space \- Coordinates of a Point in Space \- Distance between Two Points \- Section Formula \- Direction Cosines and Direction Ratios of a Line \- Equation of a Line in Space, Angle between Two Lines, Shortest Distance between Two Lines \- Plane \- Coplanarity of Two Lines \- Angle between Two Planes \- Distance of a Point from a Plane \- Angle between a Line and a Plane
------------------------------------------------------------------------------------------------------------------------------------------------
Some of the topics are obvious. Like the entire Calculus section from "Relations & Function" to "Integrals" & Vector Algebra.
And Stats and Probability.
But what about Binomial Theorem, Sequences & Series, Matrices & Determinants. And Complex Numbers.
Polynomials, Quadratics is fairly easy.
And what about he Geometry-ish section. Especially the entire Conic Sections and 3D Geometry. I am completely blanked on that. I can't remember it at all.
I can remember a fair amount of Trig and Straight Lines (Slope & distance etc). Not sure if that is needed. Trig Functions is probably important. (sine, cosine etc)
Thank very very much for even taking the time to read.
submitted by /u/BhooshanAJ [link] [comments] from math http://bit.ly/2IHjCqL from Blogger http://bit.ly/2IsYvt1
0 notes