#AlphaFold Server
Explore tagged Tumblr posts
Text
Why I Believe AlphaFold 3 is a Powerful Tool for the Future of Healthcare
Insights on a groundbreaking artificial intelligence tool for health sciences research Dear science and technology readers, Thanks for subscribing to Health Science Research By Dr Mike Broadly, where I curate important public health content. A few months ago, I wrote about AlphaFold 3, a groundbreaking AI tool that helps scientists understand protein structures, which are essential for…
#academic research#AI in healthcare#alphafold#AlphaFold 3#AlphaFold 3 represents a major leap in biological understanding#AlphaFold 3 to model proteins#AlphaFold Protein Structure Database#AlphaFold Server#Artificial Intelligence#Bioinformatics#biotechnology#Democratizing Science:#Drug discovery#future of artificial intelligence#future of science#future of technology#health#health science resarch#Health sciences research#Insights from Dr Michael Broadly#Isomorphic Labs#life lessons#Machine learning in medicine#Personalized medicine#Precision Predictions#protein resarch#Protein structure prediction#Real-World Applications#research#science
1 note
·
View note
Note
Proteins blog have you seen alphafold3? Feels like that would be something you're into
i have! and in fact this is a perfect ask to use to comment on a new improvement coming to the rest of my structures starting now!
(first, my apologies that it took me so long to start using this, and also that i haven't posted in a while. things have been crazy with assignments and finals, as well as researching master's programs since i'll be graduating soon)
sooo,, it turns out that now i can use AF3 on my computer (through AlphaFold Server)! this should give me better processing power to make some of the longer asks a little less painful, and is supposed to be better for accuracy, but honestly a lot of the main improvements aren't necessarily things that apply to this blog, such as being able to model other macromolecules, and protein interactions with ligands. it also gives me an option to add post translational modifications, which is so cool! it's really exciting and i look forward to seeing how it handles the absolute garbage i'll be throwing at it (:
letter sequence in this ask matching protein-coding amino acids:
PrteinslghaveyseenalphafldFeelslikethatwldesmethingyreint
protein guy analysis:
this is a pretty short one, so i guess it isn't the best test of AlphaFold3, but that's okay. there's really not much to say about it, since pretty much the whole thing is a long alpha helix. this doesn't show me the steps as it goes in the same way CollabFold did, and i can't find where it tells me which of the structures i can download (they give me 5 models) is considered the best, so i'm not entirely sure which one to use. for this model, 4 of them looked really similar, while one was pretty different and basically divided the helix into 2 parts. i'll try and do more research into this for later, but if anyone has any insight, please let me know! (full disclosure, for a while i had a similar issue with AF2 because at one point the order that the models showed up in the zip file changed, so some really old structures may be worse models. my apologies if i did that!)
predicted protein structure:

Structure output on AlphaFold Server website, with pLDDT and PAE scores

predicted structure (model 4)

predicted structure (model 0)
#science#biochemistry#biology#chemistry#stem#proteins#protein structure#science side of tumblr#protein asks
47 notes
·
View notes
Text
A3 Ultra VMs With NVIDIA H200 GPUs Pre-launch This Month

Strong infrastructure advancements for your future that prioritizes AI
To increase customer performance, usability, and cost-effectiveness, Google Cloud implemented improvements throughout the AI Hypercomputer stack this year. Google Cloud at the App Dev & Infrastructure Summit:
Trillium, Google’s sixth-generation TPU, is currently available for preview.
Next month, A3 Ultra VMs with NVIDIA H200 Tensor Core GPUs will be available for preview.
Google’s new, highly scalable clustering system, Hypercompute Cluster, will be accessible beginning with A3 Ultra VMs.
Based on Axion, Google’s proprietary Arm processors, C4A virtual machines (VMs) are now widely accessible
AI workload-focused additions to Titanium, Google Cloud’s host offload capability, and Jupiter, its data center network.
Google Cloud’s AI/ML-focused block storage service, Hyperdisk ML, is widely accessible.
Trillium A new era of TPU performance
Trillium A new era of TPU performance is being ushered in by TPUs, which power Google’s most sophisticated models like Gemini, well-known Google services like Maps, Photos, and Search, as well as scientific innovations like AlphaFold 2, which was just awarded a Nobel Prize! We are happy to inform that Google Cloud users can now preview Trillium, our sixth-generation TPU.
Taking advantage of NVIDIA Accelerated Computing to broaden perspectives
By fusing the best of Google Cloud’s data center, infrastructure, and software skills with the NVIDIA AI platform which is exemplified by A3 and A3 Mega VMs powered by NVIDIA H100 Tensor Core GPUs it also keeps investing in its partnership and capabilities with NVIDIA.
Google Cloud announced that the new A3 Ultra VMs featuring NVIDIA H200 Tensor Core GPUs will be available on Google Cloud starting next month.
Compared to earlier versions, A3 Ultra VMs offer a notable performance improvement. Their foundation is NVIDIA ConnectX-7 network interface cards (NICs) and servers equipped with new Titanium ML network adapter, which is tailored to provide a safe, high-performance cloud experience for AI workloads. A3 Ultra VMs provide non-blocking 3.2 Tbps of GPU-to-GPU traffic using RDMA over Converged Ethernet (RoCE) when paired with our datacenter-wide 4-way rail-aligned network.
In contrast to A3 Mega, A3 Ultra provides:
With the support of Google’s Jupiter data center network and Google Cloud’s Titanium ML network adapter, double the GPU-to-GPU networking bandwidth
With almost twice the memory capacity and 1.4 times the memory bandwidth, LLM inferencing performance can increase by up to 2 times.
Capacity to expand to tens of thousands of GPUs in a dense cluster with performance optimization for heavy workloads in HPC and AI.
Google Kubernetes Engine (GKE), which offers an open, portable, extensible, and highly scalable platform for large-scale training and AI workloads, will also offer A3 Ultra VMs.
Hypercompute Cluster: Simplify and expand clusters of AI accelerators
It’s not just about individual accelerators or virtual machines, though; when dealing with AI and HPC workloads, you have to deploy, maintain, and optimize a huge number of AI accelerators along with the networking and storage that go along with them. This may be difficult and time-consuming. For this reason, Google Cloud is introducing Hypercompute Cluster, which simplifies the provisioning of workloads and infrastructure as well as the continuous operations of AI supercomputers with tens of thousands of accelerators.
Fundamentally, Hypercompute Cluster integrates the most advanced AI infrastructure technologies from Google Cloud, enabling you to install and operate several accelerators as a single, seamless unit. You can run your most demanding AI and HPC workloads with confidence thanks to Hypercompute Cluster’s exceptional performance and resilience, which includes features like targeted workload placement, dense resource co-location with ultra-low latency networking, and sophisticated maintenance controls to reduce workload disruptions.
For dependable and repeatable deployments, you can use pre-configured and validated templates to build up a Hypercompute Cluster with just one API call. This include containerized software with orchestration (e.g., GKE, Slurm), framework and reference implementations (e.g., JAX, PyTorch, MaxText), and well-known open models like Gemma2 and Llama3. As part of the AI Hypercomputer architecture, each pre-configured template is available and has been verified for effectiveness and performance, allowing you to concentrate on business innovation.
A3 Ultra VMs will be the first Hypercompute Cluster to be made available next month.
An early look at the NVIDIA GB200 NVL72
Google Cloud is also awaiting the developments made possible by NVIDIA GB200 NVL72 GPUs, and we’ll be providing more information about this fascinating improvement soon. Here is a preview of the racks Google constructing in the meantime to deliver the NVIDIA Blackwell platform’s performance advantages to Google Cloud’s cutting-edge, environmentally friendly data centers in the early months of next year.
Redefining CPU efficiency and performance with Google Axion Processors
CPUs are a cost-effective solution for a variety of general-purpose workloads, and they are frequently utilized in combination with AI workloads to produce complicated applications, even if TPUs and GPUs are superior at specialized jobs. Google Axion Processors, its first specially made Arm-based CPUs for the data center, at Google Cloud Next ’24. Customers using Google Cloud may now benefit from C4A virtual machines, the first Axion-based VM series, which offer up to 10% better price-performance compared to the newest Arm-based instances offered by other top cloud providers.
Additionally, compared to comparable current-generation x86-based instances, C4A offers up to 60% more energy efficiency and up to 65% better price performance for general-purpose workloads such as media processing, AI inferencing applications, web and app servers, containerized microservices, open-source databases, in-memory caches, and data analytics engines.
Titanium and Jupiter Network: Making AI possible at the speed of light
Titanium, the offload technology system that supports Google’s infrastructure, has been improved to accommodate workloads related to artificial intelligence. Titanium provides greater compute and memory resources for your applications by lowering the host’s processing overhead through a combination of on-host and off-host offloads. Furthermore, although Titanium’s fundamental features can be applied to AI infrastructure, the accelerator-to-accelerator performance needs of AI workloads are distinct.
Google has released a new Titanium ML network adapter to address these demands, which incorporates and expands upon NVIDIA ConnectX-7 NICs to provide further support for virtualization, traffic encryption, and VPCs. The system offers best-in-class security and infrastructure management along with non-blocking 3.2 Tbps of GPU-to-GPU traffic across RoCE when combined with its data center’s 4-way rail-aligned network.
Google’s Jupiter optical circuit switching network fabric and its updated data center network significantly expand Titanium’s capabilities. With native 400 Gb/s link rates and a total bisection bandwidth of 13.1 Pb/s (a practical bandwidth metric that reflects how one half of the network can connect to the other), Jupiter could handle a video conversation for every person on Earth at the same time. In order to meet the increasing demands of AI computation, this enormous scale is essential.
Hyperdisk ML is widely accessible
For computing resources to continue to be effectively utilized, system-level performance maximized, and economical, high-performance storage is essential. Google launched its AI-powered block storage solution, Hyperdisk ML, in April 2024. Now widely accessible, it adds dedicated storage for AI and HPC workloads to the networking and computing advancements.
Hyperdisk ML efficiently speeds up data load times. It drives up to 11.9x faster model load time for inference workloads and up to 4.3x quicker training time for training workloads.
With 1.2 TB/s of aggregate throughput per volume, you may attach 2500 instances to the same volume. This is more than 100 times more than what big block storage competitors are giving.
Reduced accelerator idle time and increased cost efficiency are the results of shorter data load times.
Multi-zone volumes are now automatically created for your data by GKE. In addition to quicker model loading with Hyperdisk ML, this enables you to run across zones for more computing flexibility (such as lowering Spot preemption).
Developing AI’s future
Google Cloud enables companies and researchers to push the limits of AI innovation with these developments in AI infrastructure. It anticipates that this strong foundation will give rise to revolutionary new AI applications.
Read more on Govindhtech.com
#A3UltraVMs#NVIDIAH200#AI#Trillium#HypercomputeCluster#GoogleAxionProcessors#Titanium#News#Technews#Technology#Technologynews#Technologytrends#Govindhtech
2 notes
·
View notes
Text
My First Blog
Today I briefly wrote about Alphafold 3 in my blogs. I plan to share more here. Here is the link to my blog post.
2 notes
·
View notes
Link
DeepMind has once again taken a significant step in computational biology with the release of AlphaFold 3’s inference codebase, model weights, and an on-demand server. This update brings unprecedented capabilities to the already transformative Alpha #AI #ML #Automation
0 notes
Text
Perché l'intelligenza artificiale deve vedere il lato "brutto" della scienza Il potenziale rivoluzionario di AlphaFold di DeepMind nella ricerca farmacologica Una nuova versione di AlphaFold di DeepMind offre agli scienziati la capacità di prevedere le strutture delle proteine durante le interazioni con altre molecole. Questo strumento potrebbe rivoluzionare la scoperta di farmaci permettendo di anticipare modifiche che alterano la funzione delle proteine, delle strutture di DNA, RNA e altri attori cellulari. Secondo il biochimico Frank Uhlmann, questa innovazione democratizzerà la ricerca sulla biologia strutturale. Al momento, l’accesso al server AlphaFold3 è limitato per proteggere l’interesse della società spin-off di DeepMind che si occupa della scoperta di farmaci. THOR: la scoperta
0 notes
Text
Google DeepMind Drops Huge AlphaFold Update
Google Deepmind
We’re now one step closer to understanding life’s biggest mysteries—down to the molecular level. On Wednesday, Google DeepMind and Isomorphic Labs dropped a massive update to AlphaFold, their machine learning model that predicts protein structures.
Some context: Since 2018, AlphaFold has been leading the charge in predicting protein structures—a crucial step for scientists to take advantage of proteins’ unique traits. With AlphaFold 3, scientists can now model:
Highly-accurate biomolecular structures and behaviors of DNA, RNA, ligands, and ions
Chemical modifications for proteins and nucleic acids
How it works: Simply provide a list of molecules, and AlphaFold 3 can render the 3D structure and simulate interactions with other biomolecules. This update shows a staggering 50% improvement in prediction accuracy compared to previous models.
And there’s more: The new AlphaFold Server is a free, web-based tool that allows researchers to access this technology. Within the server, researchers can generate structure predictions within seconds, compared to the months or even years required for experimental methods.
The catch? The server has some restrictions about what can be modeled, particularly for drug candidate molecules.
Why it matters: These last few weeks, we’ve seen cosmic leaps of AI in the biological sciences—and AlphaFold is no exception. AlphaFold 3 is more than protein prediction modeling: It’s a disruptive tool that could revolutionize drug discovery and materials science research.
0 notes
Text
AlphaFold caused a sensation in December 2020, when it dominated a contest called the Critical Assessment of Protein Structure Prediction, or CASP. The competition, held every two years, measures progress in one of biology’s grandest challenges: determining the 3D shapes of proteins from their amino-acid sequence alone. Computer-software entries are judged against structures of the same proteins determined using experimental methods such as X-ray crystallography or cryo-electron microscopy (cryo-EM), which fire X-rays or electron beams at proteins to build up a picture of their shape.
The 2020 version of AlphaFold was the software’s second edition. It had also won the 2018 CASP, but its earlier efforts mostly weren’t good enough to stand in for experimentally determined structures, says Jumper. However, AlphaFold2’s predictions were, on average, on par with the empirical structures.
It wasn’t clear when DeepMind would make the software or its predictions widely available, so researchers used information from a public talk by Jumper, and their own insights, to develop their own AI tool, called RoseTTAFold.
Then on 15 July 2021, papers describing RoseTTAFold and AlphaFold2 appeared along with freely available, open-source code and other information needed for specialists to run their own versions of the tools. A week later, DeepMind announced that it had used AlphaFold to predict the structure of nearly every protein made by humans, as well as the entire ‘proteomes’ of 20 other widely studied organisms, such as mice and the bacterium Escherichia coli — more than 365,000 structures in total (see ‘What’s known about proteomes’). DeepMind also publicly released these to a database maintained by the EMBL’s European Bioinformatics Institute (EMBL–EBI), in Hinxton, UK. That database has since swelled to almost one million structures.
This year, DeepMind plans to release a total of more than 100 million structure predictions. That is nearly half of all known proteins — and hundreds of times more than the number of experimentally determined proteins in the Protein Data Bank (PDB) structure repository.
AlphaFold deploys deep-learning neural networks: computational architectures inspired by the brain’s neural wiring to discern patterns in data. It has been trained on hundreds of thousands of experimentally determined protein structures and sequences in the PDB and other databases. Faced with a new sequence, it first looks for related sequences in databases, which can identify amino acids that have tended to evolve together, suggesting they’re close in 3D space. The structure of existing related proteins provides another way to estimate distances between amino-acid pairs in the new sequence.
AlphaFold iterates clues from these parallel tracks back and forth as it tries to model the 3D positions of amino acids, continually updating its estimate. Specialists say the software’s application of new ideas in machine learning research seems to be what makes AlphaFold so good — in particular, its use of an AI mechanism termed ‘attention’ to determine which amino-acid connections are most salient for its task at any moment.
The network’s reliance on information about related protein sequences means that AlphaFold has some limitations. It is not designed to predict the effect of mutations, such as those that cause disease, on a protein’s shape. Nor was it trained to determine how proteins change shape in the presence of other interacting proteins, or molecules such as drugs. But its models come with scores that gauge the network’s confidence in its prediction for each amino-acid unit of a protein — and researchers are tweaking AlphaFold’s code to expand its capabilities.
By now, more than 400,000 people have used the EMBL-EBI’s AlphaFold database, according to DeepMind. There are also AlphaFold ‘power users’: researchers who’ve set up the software on their own servers or turned to cloud-based versions of AlphaFold to predict structures not in the EMBL-EBI database, or to dream up new uses for the tool.
What's next for AlphaFold and the AI protein-folding revolution
https://www.nature.com/articles/d41586-022-00997-5 Comments
#What's next for AlphaFold and the AI protein-folding revolution#nature#deepmind#alphafold2#RoseTTAFold#deep learning neural networks
1 note
·
View note
Text
DeepMind’s Protein Folding AI Is Going After Coronavirus

In late December last year, Dr. Li Wenliang began warning officials about a novel coronavirus in Wuhan, China, but was silenced by the police before tragically succumbing to the disease two months later. Meanwhile, almost simultaneously, a computer server halfway across the world started issuing worrying alerts of a potential new outbreak. The server runs software by BlueDot, a company based in San Francisco that uses AI to monitor infectious disease outbreaks for signs of early trouble.
Not enough people listened to either human expertise or AI. Then cases skyrocketed in Wuhan and spread across the world, and people had to take note.
Hindsight is 20/20, but it is remarkable that BlueDot and other machine learning-based services are beginning to catch early signs of infectious disease outbreaks—almost within the same time frame as health experts, if just for COVID-19. We often hear about AI as the next second coming of healthcare, where it can catch cases early, accelerate drug development, and personalize treatment. Yet COVID-19 is the first global pandemic to ever hold healthcare AI’s feet to the flame in a global, serious, and urgent real-world test case. In a head-to-head race, can AI actually accelerate new anti-virals or vaccines for COVID-19, something the world has never previously seen? Or will traditional biotech measures excel, in turn unveiling that AI’s hype massively outstrips reality?
MIT Technology Review recently reported an excellent piece that comprehensively looks at how AI—at its current ability level—can help us predict, diagnose, and treat novel viral threats. I’m on board with the general idea: AI’s potential is enormous.
Yet for now, don’t look to AI to help tackle COVID-19; it’s simply not ready.
That said, it is enormously helpful to see how major machine learning companies are utilizing or repositioning their technologies for tackling the crisis. People often critique AI tested in “toy cases,” or standardized, limited datasets that may have limited significance in the real world. With companies working on COVID-19, that’s no longer the case.
Ready, player, go? Here’s how one major AI player in healthtech, DeepMind, is trying to knee-cap COVID-19.
AI In “Invisible Man” Prediction
The promise of AI for accelerating medical drug discovery is almost a universally supported idea. One caveat: so far, though new drugs have been discovered using AI, no AI-based drug candidate has made it through the approval process (yet), or even demonstrated that the tech makes the whole process faster to market (yet).
In very broad strokes, AI could be enormously helpful for initial drug discovery in two main ways: one, screening through millions of chemical compounds for potential drugs in simulation tests, far faster than any human expert; two, identifying targets that new drugs can latch onto, either to reduce their impact (making people less sick), or to slow their spread among people.
For COVID-19, DeepMind is focusing on the second route. Known mostly for its algorithms that beat human players at Go, DOTA, and other games, DeepMind has nevertheless been working directly on solutions for drug discovery. Their secret sauce? AlphaFold, a deep learning system that tries to predict protein structures accurately when no similar proteins exist.
AlphaGo? Fold? Collab?
Stay with me. How a protein “looks” in 3D is essential for developing new drugs, especially for new viruses. COVID-19, for example, has really spikey proteins that jut out from its surface. Normally, human cells don’t care—they won’t let the virus inside. But COVID-19’s spikey proteins also harbor a Trojan Horse that “activates” it in certain cells with a complementary component. Lung cells have an abundance of these factors, which is why they’re susceptible to invasion.
Bottom line: if a drug is going to “fit” into a protein like a key into a lock to trigger a whole cascade of nasty reactions, then the first step is to figure out the structure of the lock. That’s what DeepMind’s AlphaFold is doing.
Thanks to a surge of global collaboration, China released the genomic blueprint of the COVID-19 virus in open-access databases, whereas others have posted online the structure of some of its proteins—either determined by experiments or through computational modeling. DeepMind is taking these data to the next level by focusing on a few understudied but potentially important proteins that could become drug or vaccine targets using machine learning.
Protein folding has been a decades-long, fundamental problem in biochemistry and drug discovery. Almost all of our existing drugs grab onto certain proteins to work, so identifying protein structure is akin to surveying the enemy landscape and figuring out best attack point simultaneously. The problem is the genetic code doesn’t translate to how proteins look. When it comes to a new virus, without predicting protein structures we’re basically fighting viruses and diseases as if they were the Invisible Man.
Traditional methods use high-tech microscopes, freezing proteins into crystal-looking entities, and other strange and expensive ways to understand their structure. Under the scope, a protein is basically a chain of chemical “letters” that wrap around itself into intricate structures—kinda like how your headphones always tangle into inconceivable structures while you’re sleeping. For DeepMind and other protein-folding efforts, the key is to predict—and then find methods to untangle—those structures.
AlphaFold stands out as a union of decades of deep learning progress, but guided by expertise from protein structure databases in the public domain. In a nutshell, AlphaFold uses genome sequences (available for COVID-19 and relatively easy to get) to predict the properties of resulting proteins that actually do the work, by looking at the “distance” of each “letter” or component that makes up a certain protein. It doesn’t predict specific sequences with special powers—such as those that bind to a cell—but offers a quick police sketch of the virus perp in sight.
There’s no doubt that AlphaFold is new to the protein-folding game. Even DeepMind itself stresses that “these structure predictions have not been experimentally verified,” but could galvanize efforts at making anti-virals and/or vaccines. For now, it’s difficult to judge how much AlphaFold will contribute to the pandemic, if at all. But by automating a critical aspect of drug discovery, it’s also en route to becoming a much larger player in the next epidemic.
Of note: all of this would not be possible without public, open-source databases of protein structures (like UniProt and the Protein Data Bank) that’s been building for decades. DeepMind’s release, posted with open access, has been lauded by fellow scientists as a way of giving back to the community.
Other Players
China’s long-time Google surrogate and AI behemoth, Baidu, is using an algorithm to predict the structure of another important biomolecule, mRNA. mRNA shuttles information from the genome to protein factories, so shoot the mRNA messenger, then the viral proteins are never born. Similarly, AI could one day potentially predict epidemics and how a virus changes over time—but it will only help if there’s enough trust to listen to the models.
Various AI companies are also making a play towards efficient diagnostics—identifying COVID-19 signs in medical scans—or other measures to support at-risk and overworked medical frontline heroes. The problem is that with any new outbreak, we don’t have enough data to train an AI, which means that they will struggle to find subtle differences in imperfect medical scans, at least for now.
So, is AI our savior? Not in this pandemic. Similar to the 2003 SARS outbreak, the best response is something that has existed for centuries: social distancing. As I mentioned previously, before COVID-19 exploded into a pandemic, science was ready to provide answers for COVID-19 as long as governments were also ready to respond. And because AI is based on scientific data and helping otherwise difficult efforts, machine learning is rapidly learning to do the same.
But perhaps ironically, COVID-19 is exposing both the best and weakest parts of AI in our current society for healthcare: great models that in theory should work, solid predictions that can be tested, but not without any recommendations without a heavy dose of skepticism. COVID-19 presents a brutal test case for AI in healthcare.
But for now, the toughest case is that of government management and what we do in response.
Note: To learn more about the Covid-19 pandemic, tune into Singularity University’s free virtual summit: Covid-19: The State & Future of Pandemics.
Image Credit: Vektor Kunst from Pixabay
0 notes
Text
DeepMind’s AlphaFold 3 Server For Molecular Life Blueprint

How AlphaFold 3 Server was constructed to predict the composition and interactions of every molecule in life
Launched in 2020, Google DeepMind’s AlphaFold 2 protein prediction model has been applied by over 2 million researchers working on cancer treatments, vaccination development, and other fields. This has allowed scholars to solve a challenge they have been working on for more than fifty years. It would have been simple for the group to sit back and relax after assisting scientists in the prediction of hundreds of millions of structures.
Google DeepMind’s AlphaFold 3 Server
Rather, they began to work on AlphaFold 3 Server. The Google DeepMind and Isomorphic Labs teams released a newer model in May that improves on their earlier models by predicting not only the structure of proteins but also the interactions and structures of all other molecules in life, such as DNA, RNA, and ligands (small molecules that bind to proteins).
Research scientist Jonas Adler of Google DeepMind claims that “looking at recent high-impact research, we made enormous progress on this decades-old open problem of protein folding with AlphaFold 2.”, researchers are moving beyond that.” “Their findings frequently dealt with more intricate topics, such as the binding of RNA or tiny molecules, which AlphaFold 2 was unable to accomplish. In order to get to the current state of biology and chemistry, we needed to be able to cover every type of biomolecule because experimental research has advanced the field.
“Everything” includes ligands, which comprise roughly 50% of all pharmaceuticals. Adrian Stecula, the research head at Isomorphic Labs, states, “We see the tremendous potential of AlphaFold 3 for rational drug design, and we’re already using it in our day-to-day work.” “All of those capabilities are unlocked by the new model, including investigating the binding of novel small molecules to novel drug targets, responding to queries like ‘How do proteins interact with DNA and RNA?,’ and examining the impact of chemical modifications on protein structure.”
An order of magnitude more potential combinations were introduced with the advent of these other molecule kinds. “There is a lot of order in proteins. There are just 20 typical amino acids, for instance,” explains Jonas. Small molecules, on the other hand, have an endless amount of space and are capable of doing almost anything. They are really varied.”
This implied that it would have been impossible to create a database with all the features. Rather, Google DeepMind have made available AlphaFold Server, a free utility that allows scientists to enter their own sequences for which AlphaFold can produce molecular complexes. It has been used by researchers to create over a million structures since its May introduction.
Lindsay Willmore, a Google DeepMind research engineer, compares it to “Google Maps for molecular complexes.” “Any user who is completely non-technical can simply copy and paste the names of their small molecules, DNA, RNA, and protein sequences, hit a button, and wait a short while.” They will be able to view and assess their forecast thanks to the release of their structure and confidence metrics.
The team greatly increased the amount of data that the newer model was trained on to include DNA, RNA, tiny molecules, and more in order to enable AlphaFold 3 to function with this far wider spectrum of biomolecules. Google is able to decide, “Let’s just train on everything that exists in this dataset that has really helped us with proteins, and let’s see how far we can get,” according to Lindsay. “And it looks like we can go a fair distance.” A change in the design for the last portion of the model that creates the structure is another significant alteration to AlphaFold 3.
AlphaFold 3 Server employs a generative model that is based on diffusion, similar to their other state-of-the-art image generation models, like Imagen. This considerably simplifies the way the model handles all the new molecule kinds, whereas AlphaFold 2 employed a complicated bespoke geometry-based module.
However, that change brought up a fresh problem: Instead of anticipating disordered sections, the diffusion model would attempt to construct an erroneous “ordered” structure with a distinct spiral form because the so-called “disordered regions” of proteins weren’t included in the training data.
The group decided to use AlphaFold 2, which is already very adept at identifying which interactions which resemble a pile of disorganised spaghetti would be disordered and which ones wouldn’t. According to Lindsay, “We were able to use those predicted structures from AlphaFold 2 as distillation training for AlphaFold 3 Server, so that AlphaFold 3 could learn to predict disorder.”
The group is excited to watch how scientists will employ AlphaFold 3 Server to progress a variety of areas, including medication development and genomics research.
“The amount of progress Google DeepMind made is amazing,” remarks Jonas. “What was extremely difficult before has now become really simple. Even though there are still many challenging issues to resolve, they are enthusiastic about AlphaFold 3 Server‘s potential to contribute to their resolution. What was once unthinkable is now achievable.
Read more on Govindhtech.com
#AlphaFold3Server#googledeepmind#AlphaFold3#AlphaFold2#GoogleMaps#generativemodels#news#technews#technology#technologynews#technologytrends#govindhtech
0 notes
Text
AlphaFold 3 Predicts All Life’s Molecules And Relations
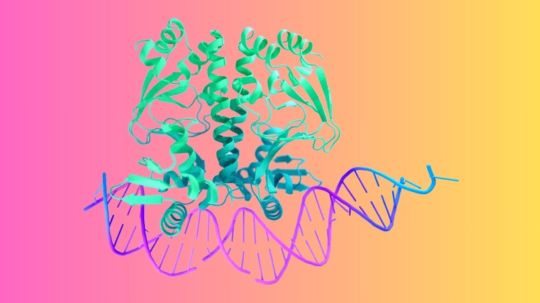
Billions of molecular machinery are found inside the cells of every plant, animal, and human. Although proteins, DNA, and other molecules make up their composition, no one component functions by itself. Google DeepMind can only begin to genuinely comprehend the workings of life by observing how these components interact among millions of possible configurations.
Google DeepMind presents AlphaFold 3, a ground-breaking model that can predict the composition and interactions of every molecule found in life with previously unheard-of precision, in a research that was published in Nature. In comparison to current prediction techniques, Google DeepMind observes at least a 50% improvement in protein interactions with other molecule types. In certain significant interaction categories, Google DeepMind has doubled prediction accuracy.
AlphaFold 3 is expected to revolutionise Google DeepMind’s comprehension of biology and drug discovery. The majority of its features are available to scientists without charge thanks to Google DeepMind’s recently released AlphaFold Server, an intuitive research tool. Isomorphic Labs is already working with pharmaceutical companies to apply AlphaFold 3 to real-world drug design difficulties in order to improve on its potential for drug design and, ultimately, discover breakthrough medicines that could change the lives of patients.
The new model from Google DeepMind expands upon the work of AlphaFold 2, which achieved a significant breakthrough in the prediction of protein structures in 2020. AlphaFold 2 has been utilised by millions of researchers worldwide to date to create breakthroughs in the creation of enzymes, cancer therapies, and vaccinations against malaria.
More than 20,000 citations have been made to AlphaFold, and its contributions to science have been acknowledged with numerous awards most recently, the Breakthrough Prize in Life Sciences. With AlphaFold 3, Google DeepMind may explore a wide range of biomolecules in addition to proteins. This breakthrough could lead to the development of more revolutionary technology, such as faster drug design and genomics research, as well as the creation of biorenewable materials and more resilient crops.
How AlphaFold 3 shows life molecules
AlphaFold 3 creates a 3D structure of molecules from a list, showing how they fit together. It simulates both small compounds, or ligands, which include a wide range of pharmaceuticals, and major macromolecules like proteins, DNA, and RNA. Moreover, AlphaFold 3 is able to simulate chemical changes to these molecules that regulate the proper operation of cells and, if disturbed, can result in illness.
The next-generation architecture and training that encompasses all molecules in life are what give AlphaFold 3 its powers. The Google DeepMind Evoformer module, a deep learning architecture that enabled AlphaFold 2’s remarkable performance, has been refined and is at the heart of the model. Following the input processing, AlphaFold 3 uses a diffusion network, similar to those in AI picture generators, to put together its predictions. After taking numerous steps, the diffusion process eventually converges on the most precise molecular structure from a starting cloud of atoms.
The molecular interactions predicted by AlphaFold 3 are more accurate than those by any other technology now in use. It is the only model that can comprehensively compute complete chemical complexes, making it unique in its ability to bring scientific discoveries together.
Lead drug discovery at Isomorphic Labs
With predictions for frequently used compounds in medications, like ligands and antibodies, which bind to proteins to alter how they interact in human health and disease, AlphaFold 3 develops capabilities for drug design.
When it comes to anticipating drug-like interactions, such as the binding of ligands and antibodies to their target proteins, AlphaFold 3 achieves previously unheard-of levels of accuracy. Without requiring structural knowledge to be input, AlphaFold 3 outperforms the best traditional approaches by 50% on the PoseBusters benchmark, becoming the first artificial intelligence system to outperform physics-based tools for biomolecular structure prediction. Antibody-protein binding prediction is essential for comprehending the human immune response and for developing novel antibodies, which are an expanding class of medicines.
Drug design for both internal projects and pharmaceutical partners is being worked on by Isomorphic Labs using AlphaFold 3 in conjunction with a corresponding suite of in-house AI models. By utilising AlphaFold 3 to aid comprehend how to approach new disease targets and provide creative approaches to pursue existing ones that were previously unattainable, Isomorphic Labs is able to expedite and enhance the success of medication design.
AlphaFold Server: Free, simple research tool
The most precise tool in the world for predicting how proteins interact with other molecules throughout the cell is Google DeepMind’s recently released AlphaFold Server. Scientists from all over the world can use it for free for non-commercial study. Biologists can use AlphaFold 3’s capabilities to simulate structures made of proteins, DNA, RNA, and a variety of ligands, ions, and chemical changes with only a few clicks.
AlphaFold Server expedites workflows and fosters additional creativity by assisting scientists in developing new ideas to test in the lab. Regardless of their level of machine learning experience or access to computational resources, researchers may easily make predictions using the Google DeepMind platform.
Approximately as long as a PhD programme, experimental protein-structure prediction can go into the hundreds of thousands of dollars. At the current rate of experimental structural biology, it would have required hundreds of millions of researcher-years to forecast hundreds of millions of structures using Google DeepMind’s previous model, AlphaFold 2.
Responsible AlphaFold 3 power sharing
Google DeepMind has collaborated with the safety and scientific communities to comprehend the wider implications of each AlphaFold release. Google DeepMind has a science-led strategy and has carried out in-depth analyses to convey the broad advantages to biology and humanity while minimising possible hazards.
Expanding upon the external consultations Google DeepMind conducted for AlphaFold 2, Google DeepMind is currently interacting with over fifty domain experts and specialised third parties from the fields of biosecurity, research, and industry to comprehend the potential hazards and capabilities of the upcoming AlphaFold models. Prior to AlphaFold 3’s release, Google DeepMind also took part in community-wide forums and conversations.
AlphaFold Server is a reflection of Google DeepMind’s continued dedication to sharing AlphaFold’s advantages, such as the 200 million protein structures in Google DeepMind’s free database. Together with EMBL-EBI and partnerships with organizations in the Global South, Google DeepMind will also be expanding its free online course AlphaFold education in order to give scientists the resources they need to speed up adoption and research, particularly in underfunded fields like food security and neglected diseases. Google DeepMind will keep collaborating with academics, policymakers, and industry leaders to responsibly develop and implement AI technology.
New AI-powered cell biology frontier
The biological world is brought to life in high resolution in AlphaFold 3. It enables researchers to view biological systems in all of their complexity, including their connections, structures, and alterations. This new perspective on life’s molecules shows how they are interconnected and aids in understanding how those connections impact biological processes including the action of medications, hormone synthesis, and DNA repair, which maintains health.
The benefits of Google DeepMind’s free AlphaFold Server and AlphaFold 3 will be felt in the way they enable researchers to explore new areas of study and answer unanswered biological issues more quickly. AlphaFold 3’s potential is only now being realised by Google DeepMind, and they are eager to see what the future brings.
Read more on govindhtech.com
#ai#aimodels#AlphaFold3#AlphaFoldServer#alphafold#AITechnology#googledeepmind#artificialintelligence#news#technews#technology#technologynews#technologytrends#govindhtech
0 notes