#DNA-Encoded Chemical Libraries
Explore tagged Tumblr posts
Text
Global DNA Encoded Libraries - A Revolutionary Approach To Drug Discovery

DNA encoded libraries are a powerful new method for drug discovery. They allow scientists to rapidly screen billions of drug-like small molecules to find new candidates for drug development.
How do they work?
DELs work by attaching short Global DNA Encoded Libraries tags to individual drug-like small molecules. Each small molecule is given a unique DNA barcode. Large libraries containing billions of these DNA-tagged molecules can then be synthesized and stored.
To screen the library, the tagged molecules are incubated with a biological target, like a protein involved in disease. Any molecules that bind to the target will be captured along with their unique DNA barcode. Scientists can then determine what molecules bound by decoding the DNA sequences. This allows high-throughput identification of potential lead compounds for drug development from enormous libraries of molecules.
Advantages Over Traditional Screening Methods
Traditional high-throughput screening methods for drug discovery analyze molecules one at a time in microplate wells. They can only test around one million compounds per day. DNA encoded libraries overcome this limitation by allowing all the molecules in a library to be screened simultaneously. Experiments can identify binders from billions of molecules in a single assay.
They also have advantages over fragment-based drug discovery methods. Fragment screens identify small chemical fragments that bind to targets, which must then be elaborated into lead compounds. They start with drug-sized molecules, so hits require less optimization.
Global Expansion Of Dels Technology
Since their development in 2015, they have revolutionized drug discovery across the pharmaceutical and academic research. Major pharmaceutical companies like Pfizer, GSK, Janssen, and Sanofi have all established it screening programs.
In 2020, Anthropic established the world's largest public DNA encoded library of over 31 billion molecules, opening up this powerful screening technology for academic and non-profit research groups globally. The library includes both commercially available compounds and novel structures synthesized in-house.
Anthropic's library has screened over 100 biological targets from research collaborators worldwide. Hits identified include leads against malaria, tuberculosis and neglected tropical diseases. The shared library model enables researchers to screen billions of molecules for a nominal fee, democratizing access to this advanced drug discovery approach.
Advancing Precision Medicine With DEL
DELs hold great promise for advancing precision medicine and developing therapeutics targeted to specific patient genomes or biomarkers. Researchers can now screen entire genomic or protease mutant libraries against the growing number of known disease-associated protein variants and mutants.
This allows high-resolution mapping of how genomic changes and mutations alter the binding profiles of drug targets - revealing opportunities for precision therapies. Combining DELs screening with multi-omics patient data also enables the discovery of biomarker-targeted drug candidates from day one of development.
Global Regulatory Acceptance And Clinical Validation
As DELs screening has matured,regulatory agencies are increasingly recognizing the approach. In 2020, the FDA approved the first new drug developed using a DNA encoded library by Astex Pharmaceuticals, called gilteritinib, for the treatment of acute myeloid leukemia.
This landmark approval demonstrated regulatory acceptance of DELs as an established drug discovery technology. It has encouraged further investment and validation efforts by pharmaceutical companies to advance hits from DNA encoded screening into clinical candidates and new medicines.
With the establishment of large shared public libraries like Anthropic’s, DNA encoded screening is becoming a powerful global resource for drug discovery. It will continue to transform both academic and industrial new drug research by massively expanding the chemical space that can be rapidly explored for novel bioactive DELs are set to play a major role in developing the medicines of tomorrow.
In DNA encoded libraries represent a revolutionary new approach to drug discovery that is being rapidly adopted globally. By enabling high-throughput screening of billions of drug-like molecules against disease targets simultaneously, they have far surpassed traditional screening methods in scale and efficiency.
Through both industrial applications and public library sharing programs, DNA encoded screening allows researchers worldwide to identify novel lead compounds that may ultimately become new medicines. They are also advancing precision medicine through unbiased exploration of genomic and patient biomarker datasets. With regulatory acceptance growing, DNA encoded libraries will continue to transform drug R&D and deliver new treatments to patients.
Get more insights on this topic: https://www.ukwebwire.com/global-dna-encoded-library-revolutionizing-drug-discovery-through-dna-encoded-chemical-libraries/
About Author:
Ravina Pandya, Content Writer, has a strong foothold in the market research industry. She specializes in writing well-researched articles from different industries, including food and beverages, information and technology, healthcare, chemical and materials, etc. (https://www.linkedin.com/in/ravina-pandya-1a3984191)
*Note: 1. Source: Coherent Market Insights, Public sources, Desk research 2. We have leveraged AI tools to mine information and compile it
#Global DNA Encoded Library#Clinical Candidates#Revolutionizing Drug Discovery#DNA-Encoded Chemical Libraries
0 notes
Text
DNA Mishaps: When the Script Gets Flipped!
DNA, the molecule that holds the blueprint of life, isn't always static. It's like a library of instructions, constantly copied and passed on. But sometimes, errors creep in, leading to changes in the genetic code known as mutations. These alterations can be small and subtle, or large and dramatic, impacting the organism in various ways.
Imagine you're writing a super important essay, and accidentally mix up the letters. Instead of "the quick brown fox jumps over the lazy dog," you end up with "the qick brown foz jmups ovetr te laxy dog." Oops! This, my friends, is kind of what happens in DNA mutations. But instead of an essay, it's the blueprint of life getting a little jumbled. Understanding these changes is crucial, as they hold the key to understanding evolution, genetic diseases, and even the potential for future therapies. Sometimes, due to mistakes during copying or exposure to things like radiation, chemicals, or even sunlight, those A, T, C, and G chemicals get swapped, added, or deleted. It's like the gremlin wrote "foz" instead of "fox."
Let's dive into the wacky world of DNA mutations
Mutations come in all shapes and sizes, classified based on the extent of the change:
Point Mutations: These are the most common, involving a single nucleotide (the building block of DNA) being substituted, deleted, or inserted. Think of these as single typos. One little DNA letter gets swapped for another. Sometimes it's harmless, like mistaking "flour" for "flower" (just add more water!). But other times, like switching "sugar" for "salt," it can completely change the outcome Point mutations can be: 1. Silent: No change in the encoded protein, like a synonym in language. 2. Missense: A different amino acid is incorporated, potentially impacting protein function. 3. Nonsense: The mutation creates a "stop codon," prematurely terminating protein production.
Insertions & Deletions: It's like adding or removing words from a sentence. These larger mutations involve adding or removing nucleotides, disrupting the reading frame and potentially causing significant functional changes.
Chromosomal Mutations: When entire segments of chromosomes are duplicated, deleted, inverted, or translocated (swapped between chromosomes), the impact can be far-reaching, affecting multiple genes and potentially leading to developmental disorders.
More Than Just a Glitch: Mutations can be beneficial, neutral, or detrimental. Some mutations are neutral, like a typo you don't even notice. But others can be like changing "hilarious" to "hairless" – they might have a big impact. Beneficial mutations, like the one enabling lactose tolerance in some humans, drive evolution. Neutral mutations have no impact, while detrimental ones can cause genetic diseases like cystic fibrosis or sickle cell anemia.
Where Do Mutations Occur? Mutations can happen in two types of cells: Germline Mutations: These occur in egg or sperm cells, meaning they get passed on to offspring, potentially impacting future generations. Somatic Mutations: These occur in body cells after conception and don't get passed on, but can contribute to diseases like cancer.
Scientists use various techniques to study mutations, from analyzing individual DNA sequences to tracking mutations across populations. This research helps us understand the causes and consequences of mutations, potentially leading to therapies for genetic diseases and even the development of new drugs.
Mutations are not errors, but rather the dynamic fuel of evolution. Thankfully, our cells have built-in proofreaders who try to catch and fix these typos. But sometimes, mutations slip through. By understanding their types, impact, and study, we gain a deeper appreciation for the intricate dance of life, where change and adaptation intertwine to create the diverse tapestry of the living world.
#life science#science#science sculpt#molecular biology#dna#mutations#genetics#somatic mutation#germline mutation#double helix#biotechnology
5 notes
·
View notes
Text
0 notes
Text
What If DNA Could Store All Human Knowledge 2025

What If DNA Could Store All Human Knowledge 2025
Imagine a future where the entire scope of human knowledge — every book, every film, every scientific discovery, and every moment of recorded history — could be encoded and stored inside something as small and essential as a strand of DNA. This idea is no longer purely science fiction. It is based on emerging scientific breakthroughs that combine the powers of biotechnology and information science. So, what would the world look like if DNA could really store all human knowledge? Let’s dive deep into the concept, its implications, and the possibilities it might unlock for the future of information, humanity, and even consciousness itself. Understanding DNA as a Data Storage Medium DNA, the molecule that carries genetic instructions in all living organisms, is incredibly dense when it comes to information storage. Just four nucleotide bases — adenine (A), thymine (T), cytosine (C), and guanine (G) — can be arranged in sequences that represent binary data (0s and 1s), much like the data stored on your phone or computer. In fact, researchers have already successfully encoded images, videos, entire books, and even operating systems into strands of synthetic DNA. For example, in a famous experiment, scientists encoded Shakespeare’s sonnets, an audio file of Martin Luther King Jr.'s “I Have a Dream” speech, and a JPEG image of the Mona Lisa into DNA and retrieved it back with nearly perfect accuracy. The Benefits of Using DNA to Store Human Knowledge Storing data in DNA comes with several major advantages over traditional digital storage systems: - 🧬 Extreme Density: DNA can store up to 215 petabytes (215 million gigabytes) per gram. - 🧬 Longevity: DNA can last for thousands of years if stored in a cool, dry place. Digital hard drives, in contrast, degrade after a few decades. - 🧬 Stability: Unlike magnetic tapes or SSDs that are prone to failure, DNA remains chemically stable for centuries. - 🧬 Universality: DNA is universal, meaning it can be read and copied by any biological system — making it a kind of "future-proof" data format. Now, imagine being able to store the entire internet inside a test tube. This is not a metaphor — it’s a real projection of future possibilities. How Would This Work in Practice? To make DNA storage practical on a global scale, a few technical challenges would need to be solved: - Encoding data into DNA involves converting binary code into sequences of A, T, C, and G. - Synthetic DNA is created using chemical processes that place these sequences in the desired order. - Reading the information back requires sequencing the DNA and decoding it into digital data. At present, this process is expensive and slow. However, rapid advances in biotechnology and AI-driven lab automation are reducing both cost and time. Within a few decades, we could see commercial DNA data storage systems as viable alternatives to cloud storage and hard drives.




Social and Scientific Implications If DNA becomes the ultimate storage device for human knowledge, it could change our society in numerous ways: 1. Revolutionizing Libraries and Archives Physical and digital libraries today consume vast amounts of space, energy, and maintenance. DNA-based libraries would require just a fraction of that space and could survive natural disasters, electromagnetic pulses, or even global internet blackouts. Imagine a tiny capsule carrying every piece of literature, every film, every academic journal — not on a server or in a vault, but in a genetic capsule you could carry in your pocket. 2. Personalized Knowledge Storage People might someday choose to carry personalized knowledge banks encoded in DNA. These could include medical records, learning materials, or even their entire family history. These capsules could be implanted subcutaneously or kept as heirlooms. 3. Integration with Human DNA (A Controversial Twist) The idea of integrating human knowledge directly into a person’s DNA is extremely controversial. But in theory, synthetic sequences could be inserted into non-coding (junk) DNA regions in human cells. This would not impact biological function but could allow for a permanent, inheritable archive of information. While this would raise significant ethical, biological, and privacy concerns, it opens the door to profound possibilities — like transmitting encyclopedic knowledge through generations. Ethical, Legal, and Privacy Concerns This kind of transformative technology doesn’t come without questions: - Who owns the DNA containing human knowledge? - Could it be hacked, corrupted, or stolen? - What if someone stores harmful, illegal, or misleading data? - Should human genomes be used as storage at all? Just like the internet needed laws, standards, and security protocols, DNA data storage will need ethical guidelines and regulatory oversight. Philosophical Questions The concept touches on deep philosophical questions as well: - What is the essence of human knowledge? - If DNA can carry all knowledge, does it bring us closer to a form of digital immortality? - Could one eventually upload parts of their consciousness, memories, or identity using DNA as a carrier? While those questions may remain speculative for now, they are no longer just the musings of science fiction writers — they are becoming real issues that future generations might confront. Potential Drawbacks and Limitations Despite the promise, several barriers still exist: - High Cost: Encoding data into DNA remains expensive and slow. - Read/Write Speeds: Accessing DNA-based data is slower than with digital drives. - Data Mutability: DNA is very stable, but in biological systems, it can mutate. This might be a concern if synthetic DNA interacts with living organisms. However, given the pace of innovation in biotechnology, machine learning, and nanotechnology, these issues may become solvable sooner than we expect. Final Thoughts Storing all human knowledge in DNA is not only feasible — it may become essential. As digital data creation continues to grow exponentially, we’re quickly reaching the physical and economic limits of traditional storage systems. DNA offers a biologically inspired solution with unmatched density, durability, and universality. So, what if DNA could store all human knowledge? The answer might be this: it would change everything — from how we preserve our past to how we shape our future. We would no longer be limited by hard drives or server farms. Instead, we could embed the legacy of humanity into the very fabric of life. And perhaps, one day, a strand of DNA floating in a glass vial could contain the entire story of civilization — all within a few microscopic coils. 📚 Explore our other futuristic topics: - What If Dreams Could Be Recorded and Played Back 2025 https://www.edgythoughts.com/what-if-dreams-could-be-recorded-and-played-back-2025 - What If Humans Could Communicate via Brain-to-Brain Networks 2025 https://www.edgythoughts.com/what-if-humans-could-communicate-via-brain-to-brain-networks-2025 🌐 For more context, visit the Wikipedia page on DNA digital data storage: https://en.wikipedia.org/wiki/DNA_digital_data_storage Read the full article
#1#2#2025-01-01t00:00:00.000+00:00#2025https://www.edgythoughts.com/what-if-dreams-could-be-recorded-and-played-back-2025#2025https://www.edgythoughts.com/what-if-humans-could-communicate-via-brain-to-brain-networks-2025#215#215000000#3#academicjournal#adenine#archive#automation#binarycode#binarydata#biologicalsystem#biotechnology#byte#capsule(fruit)#cell(biology)#chemicalprocess#chemicalsubstance#civilization#cloud#cloudstorage#communicationprotocol#computer#computerdatastorage#concept#consciousness#cost
0 notes
Text
Comprehensive Analysis and Forecast of the DNA Encoded Semiconductor Libraries Market up to 2033
Market Definition
The DNA encoded semiconductor libraries market involves the development and use of semiconductor libraries that are encoded with DNA sequences for applications in fields like drug discovery, biotechnology, and materials science. These semiconductor libraries integrate DNA-based encoding techniques with semiconductor technology, enabling the creation of vast libraries of molecules or compounds that can be screened for specific properties or interactions. The use of DNA as an encoding medium allows for the rapid generation and analysis of diverse molecular structures, which is crucial for innovations in personalized medicine, targeted therapies, and advanced material design.
To Know More @ https://www.globalinsightservices.com/reports/DNA-Encoded-Semiconductor-Libraries-Market
DNA Encoded Semiconductor Libraries Market is anticipated to expand from 4.2 billion in 2024 to 9.8 billion by 2034, growing at a CAGR of approximately 8.8%.
Market Outlook
The DNA encoded semiconductor libraries market is poised for significant growth, driven by advancements in biotechnology, semiconductor technology, and the increasing need for faster, more efficient drug discovery and material development processes. DNA encoded libraries offer a unique combination of high-throughput screening, versatility, and precision, making them invaluable tools for researchers looking to identify novel bioactive compounds, potential drug candidates, and new materials.
In the pharmaceutical and biotechnology industries, DNA encoded libraries are revolutionizing drug discovery by enabling the rapid identification of lead compounds that can be further developed into therapeutic agents. By encoding large numbers of chemical compounds on DNA strands, researchers can quickly screen vast libraries of molecules for specific biological activities, dramatically accelerating the process of drug development.
The market is also benefiting from the increasing interest in personalized medicine, as DNA encoded libraries facilitate the development of drugs that are tailored to an individual’s genetic makeup, improving the efficacy and safety of treatments. Additionally, the ability to design and synthesize new materials with specific electronic, optical, or mechanical properties through DNA encoded libraries opens up new possibilities in semiconductor and nanotechnology fields, further driving market growth.
Request the sample copy of report @ https://www.globalinsightservices.com/request-sample/GIS10578
0 notes
Text
Compound Management
Compound Management Market Size, Share, Trends: Brooks Automation Leads
Integration of Artificial Intelligence and Machine Learning in Compound Management Systems Revolutionizes Sample Tracking and Analysis
Market Overview:
The global compound management market is projected to grow at a CAGR of 14.2% from 2024 to 2031, reaching USD 724.3 million by 2031. North America dominates the market due to increasing drug discovery activities, rising demand for outsourcing compound management services, and the growing adoption of automated compound management systems. The market is expanding rapidly, driven by the increasing complexity of drug development procedures, the growing number of drug candidates in pharmaceutical pipelines, and the need for efficient storage and retrieval of chemical and biological samples. There is a significant transition to automated methods and integrated software solutions to enhance productivity and reduce human error in compound library management.
DOWNLOAD FREE SAMPLE
Market Trends:
The compound management industry is witnessing a significant shift towards intelligent systems, driven by the integration of artificial intelligence (AI) and machine learning (ML) technologies. These technologies are revolutionizing sample tracking and analysis, with companies investing in smart compound management platforms that can predict sample degradation, optimize storage conditions, and streamline retrieval processes. For example, Brooks Life Sciences' BioStore III Cryo automated storage system uses AI algorithms to predict and prevent sample integrity issues. This trend is further supported by the rising demand for predictive analytics in drug discovery.
Market Segmentation:
Chemical compounds dominate the compound management market, accounting for a significant share due to the widespread use of small molecule libraries in drug discovery and development. The segment has seen substantial progress thanks to innovations in storage technologies, sample tracking systems, and integration with high-throughput screening platforms. Efficient management of chemical compound libraries is crucial for expediting drug discovery. Advanced compound management systems allow researchers to store, retrieve, and track millions of compounds efficiently, reducing the time and cost of early-stage drug development. The growth of DNA-encoded libraries (DELs) has further increased the demand for advanced chemical compound management solutions.
Market Key Players:
Brooks Automation (Azenta Life Sciences)
Hamilton Company
TTP Labtech (SPT Labtech)
Tecan Group
Evotec
Titian Software
Contact Us:
Name: Hari Krishna
Email us: [email protected]
Website: https://aurorawaveintellects.com/
0 notes
Text
TAFAKKUR: Part 184
Recurring DNA in Genome Structure: Part 1
A genome is a data book or registry which records the past and future of living organisms. It dynamically and simultaneously stores hereditary and biological information in three different hierarchical levels belonging to three different time periods.
The first is the preservation of characteristic, long term data imprints that describes the development of an organism in the stable DNA sequences.
Second is the storage of medium term epigenetically featured data that is carried a couple of generations further down the cellular level. Epigenetic information is not stored within nucleotide sequences but in the chemical modifications of these sequences (like the methylation of repeated strings of GC dinucleotide).
Third is the storage of data generated as a result of dynamic interactions between proteins, RNA and DNA in order to adapt to the events and changes during cellular life cycle in the form of nucleoprotein or DNA-protein complexes.
The data generation and storage capacity of DNA in three different hierarchical levels and time periods demonstrates that genome plays a plethora of roles in cellular activities and heredity. Formatting of genome for its generation and storage of data is carried out via DNA sequences of various features. Genomic system is composed of repeating DNA sequences. DNA sequences (satellite) function as a marker as they repeat numerous times in various frequencies. Genome includes genomic folders similar to that of computer systems. These genomic folders, also known as the epigenetic index of genomes, are responsible for the remodeling of chromatin and the coordinated control of genomic functions. Repeating DNA sequences play a critical role in replication of genome (making a copy of DNA), dispersal of copied DNA into daughter cells and construction of support systems that enable organization of chromatins.
It is possible to better understand genomic functions in relation to examples such as memory sticks and hard drives that are used in electronic information systems. The difference between a genome as a basic data-information storage medium from a hard disc is that it can be replicated as required by its nature and these replicas can be transferred to daughter cells. Following examples could be given to illustrate that a genome gains function only when it interacts with various data processing modules in the cell.
A copy of genome is produced by cellular DNA replication system
Correct localization of each genome copy towards daughter cells is only possible when chromosome segregation system works (the centrosomes and microtubules)
The central transcription system is responsible for the copy of data from DNA to RNA. Different gene expression patterns are developed via regulation of transcription time and level with the help of transcription factors and a web of cell signalization.
Very intricately organized genomic system structures are designed through the successive combination of protein encoding sequences, signals distributed in various places and repeating DNA sequences. Formatting of genome resembles formatting of computer programs. Various repeated serial commands of computer software are used to allocate addresses to files independent of the original data contained; different computer systems use different signals and structures to manage programs. In a similar fashion, diverse living species often utilize repeating DNA sequences and chromosomal structures to organize the encoded information and to format their genomes.
Diversity and variation of repeating DNA sequences are building blocks that are constructed into different genomic system structures. Genomes of different organisms bear characteristic system morphology just like computers with various operating systems and hardware. For instance, animal cells are created as a good model to take and incorporate foreign DNA into their genomes. Genetic data transfer among organisms of the same kind is referred to as “vertical gene transfer” whereas transfers between different species, genuses and classes are called “horizontal gene transfer.” Mobile DNA sequences like transposons are very effective horizontal gene transfer agents.
Cellular differentiation and morphogenesis (formation of tissue and organ from cell) is not programmed completely in the primary structure of the DNA sequence. Components of modular programs are encoded in a flexible way and a continuous renewed and recombined arrangement is enabled when needed.
The reason behind creation of different organisms from a single genome is this utilization of such genomic structure. Metamorphosis, that is the development of different organisms like invertebrates such as a caterpillar and a butterfly, is a good example of this feature.
Two organisms from the perspective of the same genomic protein and RNA codes can be considered as two different species. Different genomic structures and repetition of sequences among different organisms are distinctive criteria for the identification of species since these features can lead to mismatch of reproductive cells, different expression patterns of genetic code sequences, and may cause ecological diversity as well. That is why repeated DNA sequences are very important in studying parental relationships. Today, microsatellite DNA as repeated DNA sequences are used to configure biological relations among individuals in forensic sciences. Plant species vary in respect to the repeated sequences in centromeres in their chromosomes; these variations are used for identification of species. Main determinants of genomic system structure are diversity, frequency, and genomic localization of repeated DNA sequences. To explain this with examples, we could say that successively repeated sequences at centromeres, telomere repetitions and transcription, packing of chromatin, repeated sequences that are spread throughout genome in charge of cellular functions like nucleus localization are the main elements of the genome system structure. Genome is a single integrated system that is controlled closely and remotely via communication webs that use repeated sequences.
While explaining the Qur’anic concept of the Manifest Record (36:12) Bediuzzaman Said Nursi, the great renovator of Islamic thought in Turkey in the twentieth century, wrote that the Manifest Record expresses one aspect of Divine knowledge that is related “more to the past and future than to the present. It is a book of Divine Destiny that contains the origins, roots, and seeds of things, rather than their flourishing forms in their visible existence” (30th Word, Second Aim).
Inspired from this view, a seed can be considered as a tiny adorned form of Divinely creative command as programs and indexes and as a determinant for those programs and indexes in the organization of an entire tree. Since the Manifest Record book, as a title of Divine knowledge and command, observes the past and the future rather than the present, the genome of a grain or a seed acts like a library and an archive in which the future and past of an organism is written.
#allah#god#prophet#Muhammad#quran#ayah#sunnah#hadith#islam#muslim#muslimah#hijab#help#revert#convert#religion#reminder#dua#salah#pray#prayer#welcome to islam#how to convert to Islam#new convert#new revert#new muslim#revert help#convert help#islam help#muslim help
1 note
·
View note
Quote
Biological information is contained in agenetic language common to all organisms Cells are the basic building blocks of organisms, but even a single cell is complex, with many internal structures and manyfunctions that depend on information. The information requiredfor a cell to function and interact with other cells—the “blueprint” for existence—is contained in the cell’s genome, the sumtotal of all the DNA molecules it contains. DNA (deoxyribonucleic acid) molecules are long sequences of four different subunits called nucleotides. The sequence of the nucleotides contains genetic information. Genes are specific segments of DNAencoding the information the cell uses to make proteins (Figure1.4). Protein molecules govern the chemical reactions withincells and form much of an organism’s structure.By analogy with a book, the nucleotides of DNA are likethe letters of an alphabet. Protein molecules are the sentences.Combinations of proteins that form structures and control biochemical processes are the paragraphs. The structures andprocesses that are organized into different systems with specifictasks (such as digestion or transport) are the chapters of thebook, and the complete book is the organism. If you were towrite out your own genome using four letters to represent thefour nucleotides, you would write more than 3 billion letters.Using the size type you are reading now, your genome wouldfill about a thousand books the size of this one. The mechanismsof evolution, including natural selection, are the authors andeditors of all the books in the library of life. 1.4 DNA Is Life’s Blueprint The instructions for life are contained inthe sequences of nucleotides in DNA molecules. Specific DNA nucleotidesequences comprise genes. The average length of a single human geneis 16,000 nucleotides. The information in each gene provides the cell withthe information it needs to manufacture molecules of a specific protein. All the cells of a multicellular organism contain the samegenome, yet different cells have different functions and formdifferent structures—contractile proteins form in muscle cells,hemoglobin in red blood cells, digestive enzymes in gut cells,and so on. Therefore, different types of cells in an organism mustexpress different parts of the genome. How cells control geneexpression in ways that enable a complex organism to developand function is a major focus of current biological research.The genome of an organism consists of thousands of genes.If the nucleotide sequence of a gene is altered, it is likely thatthe protein that gene encodes will be altered. Alterations ofthe genome are called mutations. Mutations occur spontaneously; they can also be induced by outside factors, including chemicals and radiation. Most mutations are either harmful or have no effect, but occasionally a mutation improves thefunctioning of the organism under the environmental conditions it encounters. Such beneficial mutations are the raw material of evolution and lead to adaptations. Ref: Life The Science of Biology
http://letsdefinescience.blogspot.com/2020/05/biological-information-is-contained-in.html
1 note
·
View note
Text
New platform for engineering ribosomes to 'cook new cuisines'
https://sciencespies.com/biology/new-platform-for-engineering-ribosomes-to-cook-new-cuisines/
New platform for engineering ribosomes to 'cook new cuisines'


Credit: CC0 Public Domain
Synthetic biology researchers at Northwestern University have developed a system that can rapidly create cell-free ribosomes in a test tube, then select the ribosome that can perform a certain function.
The system, called ribosome synthesis and evolution (RISE), is an important step toward using ribosomes beyond their natural capabilities. The key feature of RISE is the ability to evolve ribosomes without cell viability constraints. The result could be new ways to synthesize materials, like nylon, or therapies, like new antibiotics that could address rising antibiotic resistance.
“Ribosomes have an extraordinary capability as the protein synthesis machinery of the cell,” said Michael Jewett, Walter P. Murphy Professor of Chemical and Biological Engineering and director of the Center for Synthetic Biology at Northwestern’s McCormick School of Engineering, who led the research. “But to synthesize proteins beyond those found in nature, we have to design and modify the ribosome to work with non-natural substrates. Developing ribosomes in vitro is an important part of that system, and we are very excited to have this new capability.”
The results will be published February 28 in the journal Nature Communications.
The ribosome is like the chef of translation, cooking up the synthesis of a diverse array of biopolymers, or proteins, that enable life. Researchers have already used the ribosome’s ability to build proteins to develop new biopharmaceuticals, like insulin. But teaching ribosomes to cook “new cuisines,” or make biopolymers that are new to nature, is difficult. Since the ribosome is required for the life of the cell, there are big constraints on how it can be altered.
Jewett and his group developed the new RISE system to overcome those cell viability constraints and ultimately repurpose the ribosome in ways that have never been possible before. By building DNA that encodes for ribosome mutants, the system can make hundreds of thousands of mutant ribosomes within hours. Using magnetic beads, researchers can then select ribosomes with functions that they want. This platform sets the stage to understand the fundamental constraints of the ribosome’s active site and create new biopolymers that could transform society. Additionally, the method could potentially be used to manufacture new materials to improve soldier and police protection.
“We validated the RISE method by selecting highly active ribosomes that are resistant to the antibiotic clindamycin from a library of variants,” Jewett said. “Our hope is that others will be able to use this platform to select for ribosomes that can carry out a new function.”
With the ability to evolve ribosomes at hand, Jewett’s team has separately been trying to understand which parts of the ribosome are amenable to change. In a related paper recently published in the journal Nucleic Acids Research, the team also mapped out the nucleotides of the active site of the ribosome to find out which nucleotides could be changed without breaking the ribosome. By building and testing every possible single nucleotide mutation in the active site, 180 in total, the researchers were surprised to find that 85 percent of these nucleotides possessed some flexibility and could be altered. Additionally, the method could potentially be used to manufacture new materials to improve soldier and police protection.
“It proves to us that you can change almost every nucleotide in the active site and still get a functional ribosome. This is so exciting for synthetic biology,” Jewett said.
Last year, the researchers also published a paper in which they developed a set of design rules that guide how ribosomes can incorporate new kinds of monomers not found in nature.
Together, this collection of papers provides a comprehensive platform for transforming the ribosome into a machine that can create new kinds of therapeutics and materials.
“Right now, the ribosome is a chef that can only make certain meals,” Jewett said. “We want to create many chefs that can make many different cuisines. This is a huge step forward toward that vision.”
The research is part of the Defense Department’s Multidisciplinary University Research Initiative program, which is supported by the Army Research Office.
“This collection of results represents a truly exciting step towards harnessing and adapting the biological cellular machinery to produce non-biological polymers,” said Dawanne Poree, polymer chemistry program manager, Army Research Office, an element of US Army Combat Capabilities Development Command’s Army Research Office. “If successful, this work will, in essence, bring synthetic materials into the realm of biological functions, and potentially rendering advanced, high-performance materials capable of catalysis, molecular encoding and data storage, nanoelectronics, self-healing, among many other functions.”
Explore further
Army project may lead to new class of high-performance materials
More information: Nature Communications (2020). DOI: 10.1038/s41467-020-14705-2
Provided by Northwestern University
Citation: New platform for engineering ribosomes to ‘cook new cuisines’ (2020, February 28) retrieved 28 February 2020 from https://phys.org/news/2020-02-platform-ribosomes-cook-cuisines.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no part may be reproduced without the written permission. The content is provided for information purposes only.
#Biology
1 note
·
View note
Link
Excerpt from this Daily Kos story:
It will be widely appreciated in the readership of this site that we are collectively, in this century, heading into an ever accelerating biodiversity crisis — sometimes also given the memorable title of the Sixth Mass Extinction. The one we cause, and which we are already into .. going on right now, not at any point in the near future.
In the minds of many, as far as I can see here, this issue seems to be connected to the Climate Change issue — more narrowly understood as the CO2 and other greenhouse gas emissions issue and the climate shifts that follow from it. But this is not sufficient. Climate Change strictly seen is part of the causes. But in reality, we drive the collapse of the biosphere that sustains us not “just” by climate change. It is our generalized “way of life” that drives it — even if the CO2 molecule would not have this infrared absorption band that makes it so troublesome, we’d still be on a racing course to the wall.
At a certain point, it becomes clear that to even think about extinction in terms of individual species is to commit an error of scale. If entomologists’ most dire predictions come true, the number of species that will go extinct in the coming century will be in the millions, if not the tens of millions. Saving them one at a time is like trying to stop a tsunami with a couple of sandbags. …But to think about the coming invertebrate extinctions is to confront a different dimension of loss. So much will vanish before we even knew it was there, before we had even begun to understand it. Species aren’t just names, or points on an evolutionary tree, or abstract sequences of DNA. They encode countless millennia of complex interactions between plant and animal, soil and air. Each species carries with it behaviours we have only begun to witness, chemical tricks honed over a million generations, whole worlds of mimicry and violence, maternal care and carnal exuberance. To know that all this will disappear is like watching a library burn without being able to pick up a single book. Our role in this destruction is a kind of vandalism, against their history, and ours as well.
11 notes
·
View notes
Text

#Global DNA Encoded Library#Clinical Candidates#Revolutionizing Drug Discovery#DNA-Encoded Chemical Libraries
0 notes
Text
Top Library Science Masters, Doctorates, & PhD Degree Programs 2019+
Library science and data science are comparable diploma paths with some differences. Librarians and data professionals might contribute to the design and development of data management techniques akin to databases. In addition they assist others find archival and other assets. In so doing, they bridge the gaps between people, info and technology. For essentially the most part, those in library science are concerned with the utilization of data as soon as it reaches the institution (e.g. library). Also, on how this information is categorised, circulated and accessed utilizing various reference services, bibliographies and search instruments. Information science (IS) tends to deal with the lifecycle of data - from creation to supply. Furthermore, IS packages are excited about more than how info is collected, organized, stored, interpreted and utilized. Their concern branches out with information evaluation, retrieval, distribution and protection. Masters in Library Science programs could also be listed underneath a number of names and range by college. Master of Library Science (MLS), Master of Arts, Master of Librarianship, Master of Library and knowledge Studies (MLIS), and Master of Science are some examples. Most Masters in Library Science degrees could be accomplished in two years of full time examine and are offered as each traditional and on-line degrees. What is the GRE and is it required? Who should my letters of recommendation come from? May I submit additional letters? Letters of recommendation should come from former college members or these you've got labored with in the Food trade who can attest to your academic and professional skills. What are the applying deadlines? Generally, 5/1 for Fall admission nevertheless we'll consider purposes after these deadlines for qualified students. What elements are included in the applying analysis? GPA from the undergraduate career, GRE scores,(language exams for International college students), letters of recommendation, statement of intent, resumes. Can Science Program submit my utility before I have all the required paperwork? Admissions will hold your utility and notify us as your documents turn into available. Are there any student clubs/organizations? Food Science college students are members the Food Science Student Association. Try the Facebook page and FSA blogspot for all the most recent Food Science Student Association happenings. What are the housing choices? In our environmental science program, you may be taught to make use of the principles and theories of science to solve problems while developing an understanding of the complicated steadiness between environmental issues, politics, and the welfare of the group. The curriculum develops the talents essential to work within the environmental field or to pursue graduate school objectives, and our campus location supplies a living context for classroom study via the land, marshes, streams, and ocean of the North Shore. Environmental science focuses on a examine of the natural world. You will take courses in science supplemented by electives in geography, history, sociology, politics, and philosophy to understand the many ways wherein components like human exercise, industrial development, authorities policies, and social trends have an effect on the environment. Faculty members infuse their programs with excitement and function, guiding college students through important analysis tasks and training them to make the most of state-of-the-art laboratory and area equipment. Desirous about studying more about our environmental science program? If you're a passionate environmentalist with a desire to show, Endicott presents a novel set of advantages on your way to licensure. Learn environmental science from a fantastic college at the moment! Furthermore, the sequence alone doesn't mechanically present understanding of how every section contributes to the whole cell or organism. The overarching goal for Genomic Science program is to know how the information in DNA spells out a living cell or organism. The second data problem is to read out the genome's instructions in the proper order, time, and quantity for every gene product. The biochemical answer begins with the selective readout (transcription) of each practical segment of DNA sequence (gene) in the form of RNA, which is an in depth chemical relative of DNA. The set of RNA transcripts generated for a cell known as its transcriptome. RNA, in turn, is the direct molecular instruction for a particular protein's synthesis, achieved by the cell in a course of often called translation. Selective gene readout in the chemical type of RNA, therefore, can govern the identity and amount of proteins, which are the cell's workhorse molecules and the ultimate bodily embodiment of knowledge encoded in DNA.
1 note
·
View note
Text
DNA-encoded Compound Library in New Drugs Research
DNA-encoded chemical library technology is increasingly being used for lead compound screening to accelerate the development of new drugs. It uses DNA fragments as barcodes to record the structural information of compounds. In a compound library containing hundreds of millions of chemical molecules, each molecule is linked to a unique piece of DNA.
0 notes
Text
“DNA Makes RNA Makes Protein” Is Not the Central Dogma of Molecular Biology
From page 282 in Adam Rutherford’s A Brief History of Everyone Who Ever Lived:
“Francis Crick named the core kernel of molecular biology the ‘Central Dogma’ in 1956—the idea that DNA encodes RNA that translates into proteins. ‘Dogma’ is a term that we science types have been trying to avoid since seventeenth century, give that it means an incontrovertible belief laid down without evidence by an authority. It was puzzling that Crick should apply it here in the business of science, in an endeavor that relies exclusively on evidence and never on authority.”
Rutherford later quotes Horace Judson’s The Eight Day of Creation to claim that Crick didn’t know what “dogma” meant. In a footnote on the same page, Rutherford claims that Crick used the term “dogma” because in 1957 there was no evidence for his Central Dogma, so the use of the term was meant to emphasize that “all religious beliefs were without serious foundation.”
In his Glossary, on page 384, Rutherford repeats the claim that Crick’s Central Dogma is “DNA makes RNA makes protein.”
In scholarship, a secondary source is a document that relates or discusses information originally presented elsewhere. A secondary source contrasts with a primary source, which is the original source of the information being discussed. A primary source can be a person with direct knowledge of a situation or a document created by such a person. My wife, Dr. Mina Graur, who is an historian, has repeatedly warned me against secondary sources. Again, she was proven right.
In Adam Rutherford’s book and in hundreds of textbooks that litter my office, the Central Dogma of molecular biology is defined as the information-flow pathway from DNA to RNA to proteins. The wording is usually very catchy: “DNA makes RNA makes protein.” This dogma is attributed to Francis Crick, but none of the books in my possession provide a reference.
Most of these books also state that the dogma was refuted with the discovery of reverse transcriptase by Howard Temin and independently by David Baltimore in 1970. The fact that genetic information can flow from RNA to DNA was deemed by an unsigned editorial in Nature to be a refutation of the Central Dogma.
Well, none of these claims is true.
The first mentioning of the Central Dogma is in a very long and eerily prophetic 1958 article by Francis Crick in Symposia of the Society for Experimental Biology entitled “On protein synthesis.” The title of the symposium was “The biological replication of macromolecules.” On page 153, the Central Dogma is introduced to the world.
The Central Dogma This states that once 'information' has passed into protein it cannot get out again [italics in the original]. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein. This is by no means universally held—Sir Macfarlane Burnet, for example, does not subscribe to it—but many workers no think along these lines. As far as I know it has not been explicitly stated before.
On the NIH website, a preliminary note or sketch is posted in which Crick provides an early outline of the Central Dogma, the axiom that DNA and RNA specify protein, but that protein can never specify either. The note is dated October 1956, but this note was only made public after Crick’s death in 2004. (The original note is stored in the Wellcome Library for the History and Understanding of Medicine.) Crick first publicly elaborated on the Central Dogma at a Society for Experimental Biology Symposium entitled “The biological replication of macromolecules,” held at University College London in September 1957, and published in 1958.
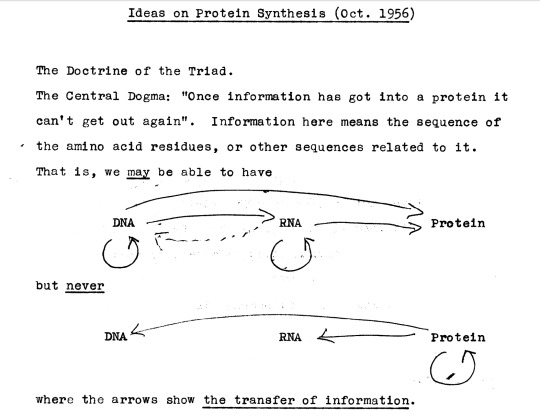
First page of a 1956 note used by Francis Crick to deliver his 1957 talk at the Society for Experimental Biology Symposium. This note became the basis for his 1958 paper that enunciates the Central Dogma
Interestingly, in the archival notes accompanying Crick sketch, the transcribed text is listed erroneously as
The Central Dogma: “Once information has got into a protein it can't get out again”. Information here means the sequence of the amino acid residues, or other sequences related to it. That is, we may be able to have
DNA --> RNA --> Protein but never DNA <-- RNA <-- Protein
where the arrows show the transfer of information.
Please note that the arrow leading from RNA to DNA does not exist in Crick’s note; it is a mistake introduced by the archiving librarian.
The figure in the note is particularly illuminating since it is clear that Crick in 1956 has already realized that the transfer of information from RNA to DNA is not a logical impossibility. The dashed line merely indicates that as of 1956, the transfer of information from RNA to DNA had not been discovered yet.
When Nature claimed in 1970 that the Central Dogma was “reversed,” Francis Crick was quite miffed, and he wrote a short paper in Nature entitled the “Central Dogma of molecular biology.” In this paper Crick dismisses the Nature editorial and writes:
This is not the first time that the Central Dogma has been misunderstood, in one way or another. In this article I explain why the term was originally introduced, its true meaning, and state why I think that, properly understood, it is still an idea of fundamental importance.
Interestingly, Crick’s definition of the Central Dogma only rarely made it into the textbooks and popular books; Nature’s mischaracterization did. A figure on page 24 of my 2016 Molecular and Genome Evolution, explains the routes of information flow according to the Central Dogma. It is a rare exception in the literature.
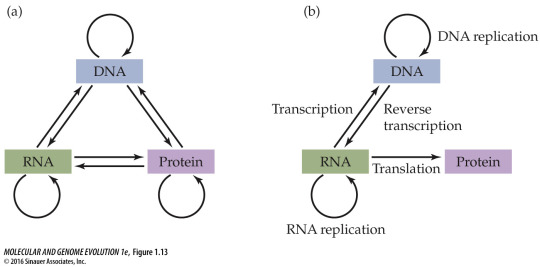
(a) Nine possible routes for information flow among DNA, RNA, and proteins. (b) The five possible routes of information flow according to Francis Crick’s Central Dogma. Only these five routes are found in nature.
The origins of “DNA makes RNA makes protein” may be older than the Central Dogma. Google Books informs me that in 1954, Edwin Chargaff is recorded as saying
“I don’t bother easily. I really don’t know. I seem to gather that the theory now is that DNA makes RNA and RNA makes protein. This may be so in special cases. I think there is some evidence that DNA makes DNA and RNA makes RNA. In fact, there is little chemical relationship at least between the total DNA of the cell and the RNA. We have looked for this but there does not seem to be any.”

Part of an exchange in a 1954 book in which we find a phrase similar to “DNA makes RNA makes protein”
I do not, however, have access to this book, and it may take some time for interlibrary loan to deliver a copy.
When did the DNA-makes-RNA-makes-protein obscenity started masquerading as the Central Dogma. It seems that it stared immediately after Crick published his 1958 paper, but the great impetus occurred after the misleading Nature editorial from 1970.
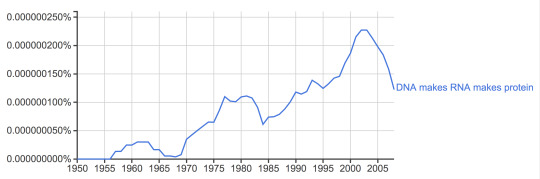
Occurrence frequency of the phrase “DNA makes RNA makes protein” in the English literature from 1950 to 2008.
Let us now return to Adam Rutherford’s book. It seems to me clear that his references to the Central Dogma are wrong. In fact, they are worse than those in Wikipedia. The Wikipedia article also provides some references to books I have not read. His description of the central dogma contain an additional element—the claim that Crick didn’t know what “dogma” meant. This claim is based on his 1979 book The Eighth Day of Creation. This claim does not seem reasonable, not only because Crick was a well educated person, but also because in his 1956 note, he seem to have considered an alternative name for what became the Central Dogma. The other name—the Doctrine of the Triad—is also derived from Christian religious nomenclature. We know that Francis Crick was deeply anti-religious, and according to Sydney Brenner, the anti-religious Francis Crick used the ecclesiastical term “dogma” as a joke. I wish Crick would have dubbed his principle, the Doctrine of the Trinity.
So, while I agree with Rutherford that Crick was a genius, I doubt that he was also an “idiot.”
Postscript 1. In 2015, two punctilious ignoramuses (Manel Esteller and Sonia Guil from the Bellvitge Biomedical Research Institute in Barcelona) published a paper in Trends in Biochemical Sciences in which they decided to “rewrite history” and invent a new definition of the Central Dogma. Accordingly, the Central Dogma was defined as “one gene gives rise to one RNA to produce one protein.” In their paper, this “new and improved” Central Dogma is thoroughly and totally refuted. Where did these people even come up with this definition?
Postscript 2. It has been suggested that prions may be construed as a refutation of the Crick’s central dogma. A prion is a protein that can fold in multiple, structurally distinct ways, at least one of which is transmissible to other prion proteins. Does this structural “contagiousness” really contradicts the central dogma. I fear not. Francis Crick defined information in the context of information flow as “the sequence of amino residues in a protein or the sequence of nucleotides in DNA or RNA.” According to this definition, a prion does not transmit protein-to-protein information.
Postscript 2. Sydney Brenner liked the DNA-Makes-RNA-Makes-Protein slogan so much that he decided to extend it to “DNA Makes RNA Makes Protein makes Money” (quoted in Masterminds: Genius, DNA & the Quest to Rewrite Life by David Ewing Duncan).
13 notes
·
View notes
Text
BMS joins forces with insitro to develop neurodegenerative treatments

insitro has landed another big biopharma partnership, signing a five-year collaboration with Bristol Myers Sqibb to develop therapies for amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
Neurodegenerative disorders such as ALS and FTD are considered a challenging therapeutic area, with no disease modifying treatments available today.
insitro uses machine-learning technology to discover novel drug targets and patient segments. Since its inception in 2018, the company has brought on several high-profile partners and investors.
Last year, the company signed its first major multi-million dollar research collaboration with Gilead Sciences to target liver disease, nonalcoholic steatohepatitis (NASH). insitro scored $15 million through the deal and could receive a further $1 billion.
Under the terms of the agreement with BMS, insitro will get $50 million in cash upfront but could be eligible for up to $2 billion if other milestones are reached. insitro’s platform, the insitro Human (ISH) platform, will be used to create induced pluripotent stem cell (iPSC) derived disease models in ALS and FTD.
The platform applies machine learning, human genetics, and functional genomics to generate predictive in vitro models that provide insights into disease progression. BMS will have the option to select targets identified by insitro and then lead clinical development. BMS said it will be responsible for regulatory submissions and commercialisation activities.
“We believe that machine learning and data generated by novel experimental platforms offer the opportunity to rethink how we discover and design novel medicines,” said Richard Hargreaves, senior vice president, head of neuroscience TRC research and early development at BMS.
Insitro recently boosted its machine-learning portfolio, with the acquisition of rival company Haystack Sciences. The private, San-Francisco based company focuses on DNA sequencing technology. It synthesises, breeds and analyses large combinatorial chemicals that are encoded by DNA sequences called DNA-encoded libraries, or DEL’s. Financial terms of the deal were not disclosed.
The post BMS joins forces with insitro to develop neurodegenerative treatments appeared first on .
from https://pharmaphorum.com/news/bms-joins-forces-with-insitro-to-develop-neurodegenerative-treatments/
0 notes