#biostatistics & bioinformatics
Explore tagged Tumblr posts
Text
alright nerds, favorite biology (or biology-adjacent) field
had to make this new poll so there's more options
#putting as many fields as i can#my bad if i left yours out!#studyblr#studying#learning#polls#tumblr polls#science#scienceblr#sciblr#biology#biological science#forensics#psychology#pathology#csi#genetics#molecular biology#paleontology#paleoblr#bioblr#college#neurobiology#neurology#pathophysiology#scientific fields#botany#bioinformatics#biostatistics#computer science
39 notes
·
View notes
Text
Common uses of bioinformatics
💡Sequence analysis Analyzing DNA and protein sequences to identify genes, regulatory regions & mutations.
💡Gene expression Analyzing RNA expression data from experiments like microarrays or RNA-seq to understand gene regulation.
💡Phylogenetics Constructing evolutionary relationships between organisms based on genetic data and genomic comparisons.
💡Molecular modeling Predicting protein structure and docking drugs to proteins using computational modeling and simulation.
💡Databases & Data mining Developing databases like GenBank to store biological data and mining it to find patterns.
💡Genomics Studying entire genomes, including sequencing and assembling genomes as well as identifying genes and genomic variations.
Follow @everythingaboutbiotech for useful posts.
#bioinformatics#genomics#proteomics#sequencing#PCR#biodata#bioIT#precisionmedicine#digitalhealth#biotech#DNA#healthtech#medtech#biostatistics#bioinformaticsjobs#BLAST#microarray#GenBank
53 notes
·
View notes
Text
fuck horoscopes and mbtis, what's your favourite R package
#programmer humor#r programming#biostatistics#bioinformatics#programming#helix talks#ggplot2#my absolute beloved#i just think its neat#trying to plot stuff with base r is suffering to me idk why#also karthik wesanderson and rcolorbrewer because heehee now your graphs can be COLOURFUL
48 notes
·
View notes
Text
"How to stay updated with the latest research in bioinformatics?
Bioinformatics is evolving rapidly, with new discoveries, algorithms, and datasets emerging every day. Whether you're a student, researcher, or professional, staying updated is crucial for growth in this interdisciplinary field. But with the overwhelming amount of information out there, how can you keep track of the latest research?
Here’s a step-by-step guide to staying informed about bioinformatics advancements:
📚 1. Follow Peer-Reviewed Journals
Leading journals publish cutting-edge bioinformatics research. Consider subscribing to:
Bioinformatics (Oxford Journals)
BMC Bioinformatics
Nucleic Acids Research (NAR)
Genome Biology
PLoS Computational Biology
Nature Biotechnology
📝 Pro Tip: Set up Google Scholar alerts for specific keywords like "machine learning in bioinformatics" or "single-cell RNA sequencing" to receive relevant papers directly in your inbox.
📰 2. Leverage Preprint Servers
Not all groundbreaking research is published in journals immediately. Many researchers upload their work to preprint servers:
bioRxiv (Preprints in biology and bioinformatics)
arXiv (Computational biology & AI in bioinformatics)
🧠 Why use preprints? They help you access fresh research before peer review, giving you a competitive edge.
💻 3. Follow Top Bioinformatics Blogs & Websites
Several platforms curate the latest developments in bioinformatics:
OMGenomics (Personal insights from bioinformatics professionals)
Bits of DNA (Exploring genomics and computational biology)
Biostars (Community-driven discussions on bioinformatics challenges)
SEQC Blog (Sequencing and bioinformatics trends)
🎙️ 4. Listen to Bioinformatics Podcasts
If you prefer learning on the go, podcasts are a great way to absorb new knowledge: 🎧 Best Bioinformatics Podcasts:
The Bioinformatics Chat
Genomics in 5 Minutes
The OmicsCast
🧑🤝🧑 5. Engage with the Bioinformatics Community
Joining discussions and interacting with experts helps you stay informed: 🔹 Reddit: r/bioinformatics, r/genomics 🔹 Twitter/X: Follow researchers and hashtags like #Bioinformatics, #ComputationalBiology 🔹 LinkedIn Groups: Bioinformatics Discussion Forum, AI in Bioinformatics 🔹 Slack & Discord: Join bioinformatics-specific communities for direct interaction
🎓 6. Take Online Courses & Webinars
Platforms like Coursera, edX, and BioPractify frequently update their courses to reflect the latest techniques in bioinformatics. Also, keep an eye out for:
Workshops by EMBL-EBI
Online tutorials from Galaxy Project & Bioconductor
📅 Tip: Many universities and conferences offer free webinars. Sign up for event notifications!
🔬 7. Attend Conferences & Hackathons
Networking at events helps you learn from researchers and industry leaders. Some key conferences include:
ISMB (Intelligent Systems for Molecular Biology)
RECOMB (Research in Computational Molecular Biology)
Genome Informatics Conference
BioHackathons (Hands-on experience with the latest tools)
🌍 Virtual & Hybrid Options: Many conferences now offer remote participation—take advantage of them!
🚀 8. Stay Hands-On with Open-Source Projects
Following GitHub repositories for bioinformatics tools and frameworks keeps you engaged with real-world applications. Some trending repositories:
Bioconductor (for R-based bioinformatics analysis)
Nextflow (for scalable data analysis workflows)
DeepVariant (AI-powered genome sequencing analysis by Google)
💡 Bonus: Contributing to open-source projects is a great way to learn while building your portfolio.
🔎 Final Thoughts
Bioinformatics is a dynamic field that blends biology, data science, and AI. Staying updated requires a multi-pronged approach—reading journals, engaging in online discussions, participating in hackathons, and continuously learning. By following these strategies, you’ll remain ahead in this ever-evolving domain.
📢 What’s your go-to method for staying updated in bioinformatics? Share your insights below! ⬇️✨
#Bioinformatics#ComputationalBiology#Genomics#MachineLearning#AIinBioinformatics#DataScience#BiotechCareers#BioinformaticsResearch#ScienceNews#Biostatistics#NextGenSequencing#BioinformaticsCommunity#ResearchTools#OpenSourceScience#GenomeAnalysis
1 note
·
View note
Text
The First Experience of Tomographic Representation of an ECG-Signal
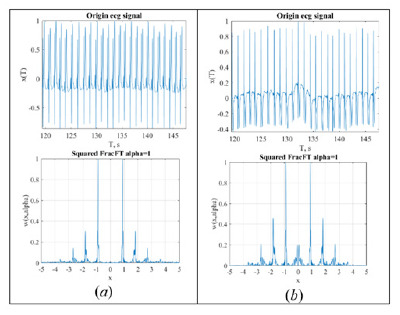
The experience for tomographic representation of ECG-signal is considered. Tomogram representation of the signal is identical to the Radon transform for processing various signals. It is shown that the tomographic representation for small parameters of transformation is identical for different leads, may reveal instability of isoelectric line and distinguish some forms of arrhythmias. This makes it possible to use only one lead for express analysis.
Read More About This Article: https://crimsonpublishers.com/oabb/fulltext/OABB.000568.php
Read More About Crimson Publishers: https://crimsonpublishers.com/oabb/index.php
#open access journals#biostatistics & bioinformatics#statistical theory#Statistical Computations#Structural genomics
0 notes
Text

🔍 Exploring Clinical SAS - Master the Art of Data in Clinical Trials! 💡📊 Unlock the power of data in the healthcare domain with our Clinical SAS Training course. Dive into the world of statistical analysis and data management for clinical trials. Ficusoft Technologies 💻🚀 📞 Contact: 94449 23024 | 94449 23025 🌐 Website: www.ficusoft.in 🔍 Key Features: 🏥 Clinical Trials Data Analysis 🖥️ Hands-on SAS Programming 🌐 CDISC Standards
#ClinicalSAS#DataManagement#HealthTech#ClinicalTrials#SASTraining#DataAnalysis#LifeSciences#HealthData#MedTech#DataScience#Biostatistics#ClinicalResearch#StatisticalAnalysis#HealthcareAnalytics#TechTraining#LearnSAS#CareerInHealthcare#DataMastery#Bioinformatics#MedResearch#ClinicalData#TechInMedicine#DataInHealthcare
0 notes
Text
#computationalbiology#bioinformatics#research#biotechnology#molecularbiology#biochemistry#microbiology#zoology#biology#biotech#immunology#science#biostatistics#lab#weirdbiotechnologists#biotechnologist#biotechnologystudents#biotechnologyjoke#animalbiotechnology#contamination#labtechnician#microbiologist#biotechlab#botany#plantscience#cellbiology#pcr#fermentation#biotechnologists#dataanalysis
0 notes
Text
NIH-Funded Postdoctoral Positions in AI/Bioinformatics for Single-Cell Omic University of Pittsburgh See the full job description on jobRxiv: https://jobrxiv.org/job/university-of-pittsburgh-27778-nih-funded-postdoctoral-positions-in-ai-bioinformatics-for-single-cell-omic/?feed_id=95174 #ai_machine_learning #Bioinformatics_and_Computational_Biology #biostatistics #ScienceJobs #hiring #research
0 notes
Text
DBT JRF 2025: Exam Date, Registration, Syllabus, and Latest Notification
The DBT JRF 2025 (Department of Biotechnology – Junior Research Fellowship) is a golden opportunity for aspirants aiming to build a research career in the field of biotechnology and life sciences. Every year, thousands of candidates apply for this prestigious fellowship exam conducted by the Department of Biotechnology under the Ministry of Science & Technology, Government of India.
In this blog, we cover everything you need to know about the DBT JRF 2025 Exam Date, notification, syllabus, registration process, and more.
What is DBT JRF 2025?
The DBT JRF 2025 is conducted to select candidates for two categories of fellowships:
Category I (JRF): For pursuing a Ph.D. in biotechnology and life sciences in universities and institutions across India.
Category II (DBT-JRF/Project): For selection in DBT-sponsored projects and programs.
The exam is officially called the Biotechnology Eligibility Test (BET), commonly known as BET DBT JRF 2025.
DBT JRF 2025 Exam Date
Candidates eagerly waiting for the DBT JRF 2025 exam date should note that the exam is generally conducted in April or May. The official exam date will be mentioned in the DBT JRF 2025 notification, expected to be released in the first quarter of 2025 on the DBT's official website.
👉 Tentative Exam Month: April 2025 👉 Mode of Exam: Computer-Based Test (CBT)
Stay tuned for the final confirmation of the dbt jrf 2025 exam date once the notification is out.
DBT JRF 2025 Notification
The DBT JRF 2025 notification is the official announcement that provides crucial information such as:
Exam date
Application start and end date
Eligibility criteria
Exam pattern
Syllabus
Reservation policy
Fellowship benefits
The DBT JRF 2025 notification is expected to be released by January or February 2025. Candidates should regularly check the official website of the Department of Biotechnology or NTA (if applicable) for updates.
DBT JRF 2025 Syllabus
The dbt jrf 2025 syllabus plays a crucial role in your preparation strategy. The BET DBT JRF 2025 syllabus is divided into two sections:
Section A: General Science, Mathematics, and Aptitude
This section includes:
General aptitude
Analytical ability
Mathematics
General biotechnology
Chemistry, Physics, and Life Sciences
Section B: Specialized Subjects in Biotechnology
This includes:
Molecular Biology
Cell Biology
Immunology
Biochemistry
Genetic Engineering
Plant and Animal Biotechnology
Microbiology
Bioinformatics
Biostatistics
Ecology and Evolution
Having a clear understanding of the dbt jrf 2025 exam syllabus will help you focus your efforts more efficiently.
✅ Download Updated DBT JRF 2025 Syllabus PDF from IFAS EdTech's official resources or the DBT portal.
️ DBT JRF 2025 Registration
The dbt jrf 2025 registration process will be entirely online. Once the DBT JRF 2025 notification is released, a registration link will be activated on the official website.
Key Documents Required:
Scanned photograph and signature
Valid ID proof
Educational certificates
Caste/Category certificate (if applicable)
How to Apply for DBT JRF 2025:
Visit the official DBT or NTA website.
Click on the DBT JRF 2025 registration link.
Fill in your details correctly.
Upload required documents.
Pay the registration fee.
Submit and download the confirmation page.
DBT JRF 2025 Exam Pattern
Understanding the exam pattern is crucial for effective preparation. The BET DBT JRF 2025 will consist of two sections: Section A and Section B.
Section A includes 50 questions based on General Science and Aptitude. This section carries a total of 150 marks, with each incorrect answer resulting in a deduction of 1 mark due to negative marking.
Section B contains 150 subject-specific questions related to Biotechnology. This section also carries 150 marks, and similar to Section A, there is a negative marking of 1 mark for every wrong answer.
Candidates are allowed to attempt a maximum of 100 questions in total from both sections combined. The final score obtained in the exam will determine whether a candidate qualifies for Category I (JRF) or Category II (Project-based positions).
Who Should Apply for DBT JRF 2025?
The DBT JRF 2025 exam is ideal for students who:
Hold a postgraduate degree in Biotechnology or related fields (M.Sc./M.Tech).
Are passionate about pursuing a Ph.D. in Biotechnology or Life Sciences.
Wish to be a part of India’s premier research programs.
Eligibility: Candidates must have a degree in Biotechnology or related discipline with a minimum qualifying percentage. Final-year students may also apply.
Why Choose IFAS for DBT JRF 2025 Preparation?
IFAS is India’s No.1 coaching institute for CSIR NET, GATE, IIT JAM, and DBT JRF 2025. Our BET DBT JRF 2025 online coaching program includes:
Expert faculty from IITs, IISc, and top research institutes
Structured video lectures covering the full dbt jrf 2025 syllabus
Topic-wise tests and full-length mock exams
Live doubt-solving sessions
Printed study material and e-books
Performance tracking with detailed analysis
Enroll in IFAS DBT JRF 2025 Online Coaching to give your preparation the edge it needs.
Tips to Crack DBT JRF 2025
Start early with a clear study plan.
Focus on high-weightage topics from the dbt jrf 2025 syllabus.
Practice with previous year papers and mock tests.
Revise regularly.
Follow current updates for the dbt jrf 2025 exam date and notification.
The DBT JRF 2025 is your gateway to a rewarding research career in biotechnology. With a well-planned strategy, a solid understanding of the dbt jrf 2025 syllabus, and timely registration, you can boost your chances of success.
Stay updated with the latest DBT JRF 2025 notification, exam date, and registration date to avoid missing out. Begin your preparation early with IFAS and take a confident step toward achieving your Ph.D. dreams.
0 notes
Text
The Growing CRO Industry in Hyderabad
Hyderabad emerged rapidly as major hub for Contract Research Organizations playing crucial role in clinical research globally nowadays. Pharmaceutical firms swiftly flock towards metropolitan areas boasting solid infrastructure skilled workers under favorable regulatory frameworks daily. Rapid expansion occurred in Hyderabad sector's CRO making it major player within global clinical research industry very rapidly nowadays.
What is a CRO?
A Contract Research Organization provides support for pharmaceutical biotechnology and medical device industries via outsourced research services somehow. Services span preclinical research and clinical trials via regulatory submissions through post-marketing studies somehow. Companies rely heavily on chief research officers for streamlining drug development processes rapidly and accelerating time-to-market. Established CROs in Hyderabad provide pharmaceutical firms with superb research facilities and remarkably skilled personnel onsite daily.
Why Hyderabad is a Preferred Location for CROs
Multiple elements fuel Hyderabad's ascent as major CRO hub with strong pharma industry led by Dr. Reddy's Laboratories nearby. Hyderabad boasts a substantial talent base of highly skilled professionals specializing in pharmacovigilance and bioinformatics fields. CROs in Hyderabad generally follow pretty strict global standards like ICH-GCP USFDA and EMA guidelines every day. Research services provided by a CRO in Hyderabad are pretty darn affordable somehow without sacrificing quality apparently. Hyderabad becomes pretty attractive somehow as destination for outsourcing clinical research apparently due its numerous advantages..
Key Services Offered by CROs in Hyderabad
CRO in Hyderabad offer myriad services for pharmaceutical and biotechnology firms across several disciplines daily.
Clinical Trials –Researchers undertake multifaceted clinical trials evaluating drug safety amidst efficacy assessments rapidly.
Bioanalytical Studies – Laboratory analysis gets done for weird pharmacokinetic drug stuff.
Regulatory Consulting –Facilitating regulatory submissions and tricky approvals under various international guidelines somehow becomes incredibly daunting.
Data Management & Biostatistics – Accurate data gets collected and analyzed painstakingly for clinical trials in a highly meticulous manner.
Pharmacovigilance –Monitoring and managing adverse drug reactions to ensure patient safety.
Companies partnering with a CRO in Hyderabad get pretty extensive support for research endeavors from these services basically.
Leading CROs in Hyderabad
Multiple seasoned CROs function within Hyderabad beneath vibrant skyscrapers and alongside bustling streets thereby bolstering its stature. Notable CROs operating out of Hyderabad are scattered throughout city limits somehow.
Syngene International
Parexel International
Cliniteq
Veeda Clinical Research
GVK Bio (Aragen Life Sciences)
These organizations provide top-notch research services thereby making them favored partners amongst pharmaceutical companies globally.
Challenges Faced by CROs in Hyderabad
CRO in Hyderabad sector faces numerous challenges rather suddenly due its fairly rapid expansion somehow.
Regulatory Complexities –Evolving regulatory requirements globally and locally necessitate fairly rapid adaptation somehow.
Talent Retention –Skilled professionals are highly sought after resulting in frequent workforce turnover.
Increasing Competition –Numerous CROs emerge and companies stay competitive by constantly innovating in highly competitive environments.
Sustaining Hyderabad's growth as a major CRO hub utterly depends on overcoming these daunting economic obstacles somehow.
Conclusion: Hyderabad’s Role in Global Clinical Research
Hyderabad will reportedly play a substantially larger role in shaping future clinical research and pharmaceutical innovation very soon. Companies partnering with a CRO in Hyderabad gain access to world-class research facilities skilled professionals and cost-effective solutions readily available overseas. Hyderabad remains a prime spot for outsourced clinical work due to ongoing investment and rapid tech advancements somehow.

0 notes
Text
Fwd: Graduate position: CzechAcadSci.PlantHerbivoreDiversity
Begin forwarded message: > From: [email protected] > Subject: Graduate position: CzechAcadSci.PlantHerbivoreDiversity > Date: 8 November 2024 at 05:59:33 GMT > To: [email protected] > > > Graduate position: Biology.Centre.CZ.Global.trends.phytochemical.diversity > > PhD Studentship in Plant-Herbivore Ecology > Global trends in phytochemical diversity and plant-herbivore interactions > > We are seeking enthusiastic candidates to join the prestigious JUNIOR > STAR project funded by the Czech Science Foundation, aimed at exploring > the global drivers of phytochemical diversity. This project investigates > the fascinating diversity of plant metabolites by analysing how various > aspects of chemical diversity correlate with biotic and abiotic stress > factors. This highly interdisciplinary research combines fieldwork > in diverse locations worldwide (Europe, USA, Panama, Cameroon, Japan) > with advanced laboratory work using advanced techniques in metabolomics, > biostatistics, and bioinformatics. Through these methods, the successful > candidate will help reveal global trends in plant chemical strategies > and their effects on multitrophic interactions and chemical communication > among plants, insect herbivores, and their natural enemies. > > We are looking for candidates that have > > * A MSc degree (non-negotiable requirement for PhD program > eligibility) > * A strong interest in chemical and community ecology of plants > and insects > * Ability to work in demanding field conditions > * Basic experience working in molecular biology or analytical > chemistry laboratories > * Excellent biostatistical skills > * Experience in bioinformatics (optional) > * Fluent spoken and written English > * Ability to work independently > * A driver's license (optional but highly recommended) > > The successful applicant will join the Ecology Department at the Institute > of Entomology, Biology Center of the Czech Academy of Science and the > Zoology Department of the University of South Bohemia. The PhD study will > be supervised by Dr. Martin Volf, leader of the Evolutionary Ecology > team. The candidate will live in Ceske Budejovice (Czech Republic) > where the studies will take place and conduct fieldwork and research > stays in Europe and overseas. Our department is a diverse, international > team studying ecology, evolution and biogeography, and a world-class > centre for interaction network research with regular publications in > leading journals. > > The deadline for applications is December 9th 2024. The best candidates > will be interviewed in mid-December. The successful applicant is expected > to start on April 1st 2025 (later start date negotiable). This five-year > project offers a scholarship fully covering living expenses in the Czech > Republic for the full standard duration of the PhD studies. Applicants > from all countries are eligible. To apply, please send a CV, contact > details for three references, and a cover letter stating qualifications, > previous work and motivation to Dr. Martin Volf ([email protected]) > where you can also send any queries. > > External Links > Volf lab: https://ift.tt/nf5j21I > Czech Academy of Science : https://ift.tt/O1UA9ZI > Zoology Department of the University of South Bohemia: > https://ift.tt/RDyFu7n > Ceske Budejovice : > https://ift.tt/P2J5Hlz > > Volf Martin
0 notes
Text
im going to beat r studio to death with a hammer
#bioinformatics#biostatistics#statistics#r programming#r studio#programming#helix talks#call me my r scripts bc im full of unhandled issues /j
36 notes
·
View notes
Text
Managing Pandemic Disasters: A Resource-Based Analysis

Healthcare crises are well-known phenomena that occur and are highly disruptive. Consider, for example, Covid19 and other pandemics, wars, multi-casualty natural disasters, industrial disasters, etc. Healthcare systems in Brazil, Spain, Italy, and more around the world collapsed due to the lack of a clear methodology for handling such crises. Following a largescale value creation project with a large medical center we developed a resource-based methodology to cope with three healthcare crisis scenarios: “Peace”, “War” and “Tsunami”.
We identified three load/capacity scenarios for hospitals: “Peace”, “War” and “Tsunami”. The Peacetime scenario is the normal overload situation in a hospital. Figure 1 during “Peace” times, hospitals are usually 10%-20% short in resources. To overcome bottlenecks, managers apply evolutionary methods such as constraint management [1], the complete kit concept, etc. This is the normal condition in hospitals where there are fluctuations in supply and demand. Bottlenecks in this situation are typically medical crews: senior physicians and experienced nurses.
Read More About This Article: https://crimsonpublishers.com/oabb/fulltext/OABB.000567.php
Read More About Crimson Publishers: https://crimsonpublishers.com/oabb/index.php
0 notes
Text
A new computational model can predict antibody structures more accurately
New Post has been published on https://sunalei.org/news/a-new-computational-model-can-predict-antibody-structures-more-accurately/
A new computational model can predict antibody structures more accurately

By adapting artificial intelligence models known as large language models, researchers have made great progress in their ability to predict a protein’s structure from its sequence. However, this approach hasn’t been as successful for antibodies, in part because of the hypervariability seen in this type of protein.
To overcome that limitation, MIT researchers have developed a computational technique that allows large language models to predict antibody structures more accurately. Their work could enable researchers to sift through millions of possible antibodies to identify those that could be used to treat SARS-CoV-2 and other infectious diseases.
“Our method allows us to scale, whereas others do not, to the point where we can actually find a few needles in the haystack,” says Bonnie Berger, the Simons Professor of Mathematics, the head of the Computation and Biology group in MIT’s Computer Science and Artificial Intelligence Laboratory (CSAIL), and one of the senior authors of the new study. “If we could help to stop drug companies from going into clinical trials with the wrong thing, it would really save a lot of money.”
The technique, which focuses on modeling the hypervariable regions of antibodies, also holds potential for analyzing entire antibody repertoires from individual people. This could be useful for studying the immune response of people who are super responders to diseases such as HIV, to help figure out why their antibodies fend off the virus so effectively.
Bryan Bryson, an associate professor of biological engineering at MIT and a member of the Ragon Institute of MGH, MIT, and Harvard, is also a senior author of the paper, which appears this week in the Proceedings of the National Academy of Sciences. Rohit Singh, a former CSAIL research scientist who is now an assistant professor of biostatistics and bioinformatics and cell biology at Duke University, and Chiho Im ’22 are the lead authors of the paper. Researchers from Sanofi and ETH Zurich also contributed to the research.
Modeling hypervariability
Proteins consist of long chains of amino acids, which can fold into an enormous number of possible structures. In recent years, predicting these structures has become much easier to do, using artificial intelligence programs such as AlphaFold. Many of these programs, such as ESMFold and OmegaFold, are based on large language models, which were originally developed to analyze vast amounts of text, allowing them to learn to predict the next word in a sequence. This same approach can work for protein sequences — by learning which protein structures are most likely to be formed from different patterns of amino acids.
However, this technique doesn’t always work on antibodies, especially on a segment of the antibody known as the hypervariable region. Antibodies usually have a Y-shaped structure, and these hypervariable regions are located in the tips of the Y, where they detect and bind to foreign proteins, also known as antigens. The bottom part of the Y provides structural support and helps antibodies to interact with immune cells.
Hypervariable regions vary in length but usually contain fewer than 40 amino acids. It has been estimated that the human immune system can produce up to 1 quintillion different antibodies by changing the sequence of these amino acids, helping to ensure that the body can respond to a huge variety of potential antigens. Those sequences aren’t evolutionarily constrained the same way that other protein sequences are, so it’s difficult for large language models to learn to predict their structures accurately.
“Part of the reason why language models can predict protein structure well is that evolution constrains these sequences in ways in which the model can decipher what those constraints would have meant,” Singh says. “It’s similar to learning the rules of grammar by looking at the context of words in a sentence, allowing you to figure out what it means.”
To model those hypervariable regions, the researchers created two modules that build on existing protein language models. One of these modules was trained on hypervariable sequences from about 3,000 antibody structures found in the Protein Data Bank (PDB), allowing it to learn which sequences tend to generate similar structures. The other module was trained on data that correlates about 3,700 antibody sequences to how strongly they bind three different antigens.
The resulting computational model, known as AbMap, can predict antibody structures and binding strength based on their amino acid sequences. To demonstrate the usefulness of this model, the researchers used it to predict antibody structures that would strongly neutralize the spike protein of the SARS-CoV-2 virus.
The researchers started with a set of antibodies that had been predicted to bind to this target, then generated millions of variants by changing the hypervariable regions. Their model was able to identify antibody structures that would be the most successful, much more accurately than traditional protein-structure models based on large language models.
Then, the researchers took the additional step of clustering the antibodies into groups that had similar structures. They chose antibodies from each of these clusters to test experimentally, working with researchers at Sanofi. Those experiments found that 82 percent of these antibodies had better binding strength than the original antibodies that went into the model.
Identifying a variety of good candidates early in the development process could help drug companies avoid spending a lot of money on testing candidates that end up failing later on, the researchers say.
“They don’t want to put all their eggs in one basket,” Singh says. “They don’t want to say, I’m going to take this one antibody and take it through preclinical trials, and then it turns out to be toxic. They would rather have a set of good possibilities and move all of them through, so that they have some choices if one goes wrong.”
Comparing antibodies
Using this technique, researchers could also try to answer some longstanding questions about why different people respond to infection differently. For example, why do some people develop much more severe forms of Covid, and why do some people who are exposed to HIV never become infected?
Scientists have been trying to answer those questions by performing single-cell RNA sequencing of immune cells from individuals and comparing them — a process known as antibody repertoire analysis. Previous work has shown that antibody repertoires from two different people may overlap as little as 10 percent. However, sequencing doesn’t offer as comprehensive a picture of antibody performance as structural information, because two antibodies that have different sequences may have similar structures and functions.
The new model can help to solve that problem by quickly generating structures for all of the antibodies found in an individual. In this study, the researchers showed that when structure is taken into account, there is much more overlap between individuals than the 10 percent seen in sequence comparisons. They now plan to further investigate how these structures may contribute to the body’s overall immune response against a particular pathogen.
“This is where a language model fits in very beautifully because it has the scalability of sequence-based analysis, but it approaches the accuracy of structure-based analysis,” Singh says.
The research was funded by Sanofi and the Abdul Latif Jameel Clinic for Machine Learning in Health.
0 notes
Text
Tumor Heterogeneity, Tumor Evolution, and Emerging Cancer Genomics National Cancer Institute, National Institutes of Health ????Postdoc Opportunity! Join our cutting-edge research team at NCI DCEG! We’re looking for talented individuals to explore cancer genomics. See the full job description on jobRxiv: https://jobrxiv.org/job/national-cancer-institute-national-institutes-of-health-27778-tumor-heterogeneity-tumor-evolution-and-emerging-cancer-genomics-2/?feed_id=91113 #bioinformatics #cancer #cancer_genomics #computational_biology #tumor_evolution #ScienceJobs #hiring #research
0 notes
Text
Crack CSIR NET Life Science 2025: You’re Ultimate Guide to Success
The CSIR NET Life Science exam is one of the most prestigious competitive exams in India for candidates aiming to qualify for Junior Research Fellowship (JRF) and Lectureship positions. Conducted by the National Testing Agency (NTA) on behalf of the Council of Scientific and Industrial Research (CSIR), this exam tests a candidate’s expertise in the field of biological sciences. If you are preparing for the upcoming CSIR NET Life Science 2025, this guide will help you with all the essential details, preparation strategies, syllabus breakdown, and expert tips to secure a top rank.
Why Appear for CSIR NET Life Science?
JRF and Lectureship Opportunities: Qualifying for JRF opens the gateway to research opportunities in top institutes, while Lectureship (LS) qualification helps you pursue a career in teaching.
Better Career Prospects: CSIR NET qualification is highly regarded in academia, research, and industry, offering excellent career growth.
Scholarship & Fellowships: Candidates qualifying for JRF receive a monthly stipend of ₹31,000 along with additional benefits.
Higher Studies: Many prestigious institutes require CSIR NET scores for Ph.D. admissions.
CSIR NET Life Science 2025: Important Exam Details
Exam Conducting Body: National Testing Agency (NTA)
Mode of Exam: Computer-Based Test (CBT)
Exam Duration: 3 Hours
Total Marks: 200
Language: English & Hindi
Marking Scheme:
Part A: General Aptitude (30 Marks)
Part B: Subject-Specific Questions (70 Marks)
Part C: Higher-Order & Analytical Questions (100 Marks)
Negative Marking: Yes (0.25 marks deducted in Part A & B)
CSIR NET Life Science 2025 Syllabus Overview
The CSIR NET Life Science syllabus is vast, covering various aspects of biological sciences. Below is the subject-wise breakdown:
1. Molecules and Their Interactions
Biomolecules: Proteins, Carbohydrates, Lipids, Nucleic Acids
Enzyme Structure & Function
Cell Membrane and Transport
2. Cellular Organization
Cell Cycle & Cell Division
Cellular Communication
Cell Signaling & Signal Transduction
3. Fundamental Processes
DNA Replication, Repair, and Recombination
Transcription & Translation
Gene Expression Regulation
4. Developmental Biology
Early Embryonic Development
Morphogenesis & Organogenesis
Stem Cells and Regenerative Biology
5. System Physiology
Animal & Plant Physiology
Neurobiology
Immunology
6. Inheritance Biology
Mendelian and Non-Mendelian Inheritance
Genetic Mapping & Epigenetics
7. Evolution & Ecology
Theories of Evolution
Population Genetics
Biodiversity & Conservation Biology
8. Methods in Biology
Microscopy & Imaging Techniques
Molecular Biology Techniques (PCR, ELISA, Electrophoresis)
Biostatistics & Bioinformatics
Best Books for CSIR NET Life Science Preparation
Molecular Biology of the Cell – Alberts
Lehninger Principles of Biochemistry – Nelson & Cox
Genetics: A Conceptual Approach – Benjamin A. Pierce
Developmental Biology – Gilbert
Ecology: The Economy of Nature – Robert E. Ricklefs
Immunology – Kuby
CSIR NET Life Science by Pathfinder – Comprehensive Practice Book
Top Preparation Strategies for CSIR NET Life Science 2025
1. Understand the Exam Pattern & Syllabus
Before diving into preparation, thoroughly go through the exam pattern and syllabus. Identify the important topics and allocate time accordingly.
2. Create a Study Plan
A well-structured study plan will help you cover the entire syllabus effectively. Divide topics into weekly and daily goals, keeping ample time for revision and mock tests.
3. Strengthen Your Basics
A strong grasp of basic concepts is crucial for solving analytical and application-based questions. Focus on fundamental topics before moving to advanced sections.
4. Practice Previous Year Question Papers
Solving previous year question papers will familiarize you with the exam pattern, difficulty level, and commonly asked questions. Aim to solve at least 10 years’ worth of papers.
5. Attempt Mock Tests Regularly
Mock tests help in time management and improve your accuracy. Analyze your performance and identify areas that need improvement.
6. Focus on High-Weightage Topics
Some topics like Molecular Biology, Genetics, and Cell Biology have higher weightage in the exam. Prioritize these sections while preparing.
7. Make Concise Notes
Prepare short notes for quick revision. Highlight important formulas, concepts, and mnemonics to aid retention.
8. Join Online Coaching & Study Groups
Enrolling in CSIR NET Life Science Online Coaching can provide structured learning, expert guidance, and doubt-clearing sessions. Join discussion groups on Telegram, WhatsApp, and Facebook to stay updated.
Time Management Tips for CSIR NET Life Science
Divide Time Wisely: Allocate 1-2 hours daily for theory, 1 hour for revision, and 1-2 hours for solving MCQs.
Use Pomodoro Technique: Study for 50 minutes followed by a 10-minute break.
Don’t Skip General Aptitude: Many students ignore Part A, but it can help boost your score.
Revise Daily: Regular revision is key to long-term retention.
CSIR NET Life Science 2025 Exam Day Strategy
Reach the center early to avoid last-minute stress.
Read the instructions carefully before starting.
Attempt easy questions first to gain confidence.
Manage time efficiently—don’t get stuck on difficult questions.
Avoid negative marking by double-checking answers.
The CSIR NET Life Science 2025 exam requires dedication, strategic preparation, and regular practice. Stay motivated, follow a disciplined study plan, and use the right resources to maximize your chances of success.
Best of luck with your CSIR NET Life Science preparation!
0 notes