#Ernest Fraenkel
Text
Review of "COUP DE DÉS (COLLECTION)"

View On WordPress
#Annette Gilbert#Arnaud Esquerre#Aurélie Noury#Benjamin Lord#Center for Book Arts#Craig Dworkin#Dan Graham#Eric Zboya#Ernest Fraenkel#Jérémie Bennequin#Klara Vith#Klaus Scherübel#Luc Boltanski#Marcel Broodthaers#Mario Diacono#Michael Maranda#Michalis Pichler#Raffaella della Olga#Rainier Lericolais#Richard Nash#Ryōko Sekiguchi#Sam Sampson#Sammy Engramer#Stéphane Mallarmé
0 notes
Text
Epigenomic analysis sheds light on risk factors for ALS
New Post has been published on https://sunalei.org/news/epigenomic-analysis-sheds-light-on-risk-factors-for-als/
Epigenomic analysis sheds light on risk factors for ALS
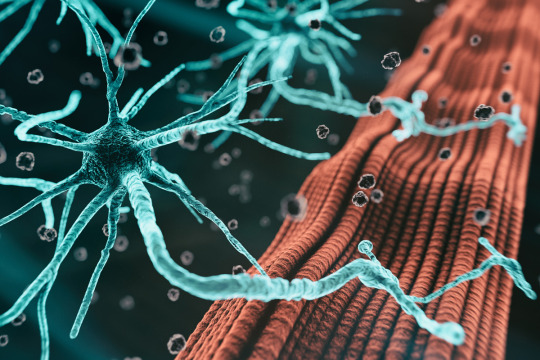
For most patients, it’s unknown exactly what causes amyotrophic lateral sclerosis (ALS), a disease characterized by degeneration of motor neurons that impairs muscle control and eventually leads to death.
Studies have identified certain genes that confer a higher risk of the disease, but scientists believe there are many more genetic risk factors that have yet to be discovered. One reason why these drivers have been hard to find is that some are found in very few patients, making it hard to pick them out without a very large sample of patients. Additionally, some of the risk may be driven by epigenomic factors, rather than mutations in protein-coding genes.
Working with the Answer ALS consortium, a team of MIT researchers has analyzed epigenetic modifications — tags that determine which genes are turned on in a cell — in motor neurons derived from induced pluripotent stem (IPS) cells from 380 ALS patients.
This analysis revealed a strong differential signal associated with a known subtype of ALS, and about 30 locations with modifications that appear to be linked to rates of disease progression in ALS patients. The findings may help scientists develop new treatments that are targeted to patients with certain genetic risk factors.
“If the root causes are different for all these different versions of the disease, the drugs will be very different and the signals in IPS cells will be very different,” says Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering and the senior author of the study. “We may get to a point in a decade or so where we don’t even think of ALS as one disease, where there are drugs that are treating specific types of ALS that only work for one group of patients and not for another.”
MIT postdoc Stanislav Tsitkov is the lead author of the paper, which appears today in Nature Communications.
Finding risk factors
ALS is a rare disease that is estimated to affect about 30,000 people in the United States. One of the challenges in studying the disease is that while genetic variants are believed to account for about 50 percent of ALS risk (with environmental factors making up the rest), most of the variants that contribute to that risk have not been identified.
Similar to Alzheimer’s disease, there may be a large number of genetic variants that can confer risk, but each individual patient may carry only a small number of those. This makes it difficult to identify the risk factors unless scientists have a very large population of patients to analyze.
“Because we expect the disease to be heterogeneous, you need to have large numbers of patients before you can pick up on signals like this. To really be able to classify the subtypes of disease, we’re going to need to look at a lot of people,” Fraenkel says.
About 10 years ago, the Answer ALS consortium began to collect large numbers of patient samples, which could allow for larger-scale studies that might reveal some of the genetic drivers of the disease. From blood samples, researchers can create induced pluripotent stem cells and then induce them to differentiate into motor neurons, the cells most affected by ALS.
“We don’t think all ALS patients are going to be the same, just like all cancers are not the same. And the goal is being able to find drivers of the disease that could be therapeutic targets,” Fraenkel says.
In this study, Fraenkel and his colleagues wanted to see if patient-derived cells could offer any information about molecular differences that are relevant to ALS. They focused on epigenomic modifications, using a method called ATAC-seq to measure chromatin density across the genome of each cell. Chromatin is a complex of DNA and proteins that determines which genes are accessible to be transcribed by the cell, depending on how densely packed the chromatin is.
In data that were collected and analyzed over several years, the researchers did not find any global signal that clearly differentiated the 380 ALS patients in their study from 80 healthy control subjects. However, they did find a strong differential signal associated with a subtype of ALS, characterized by a genetic mutation in the C9orf72 gene.
Additionally, they identified about 30 regions that were associated with slower rates of disease progression in ALS patients. Many of these regions are located near genes related to the cellular inflammatory response; interestingly, several of the identified genes have also been implicated in other neurodegenerative diseases, such as Parkinson’s disease.
“You can use a small number of these epigenomic regions and look at the intensity of the signal there, and predict how quickly someone’s disease will progress. That really validates the hypothesis that the epigenomics can be used as a filter to better understand the contribution of the person’s genome,” Fraenkel says.
“By harnessing the very large number of participant samples and extensive data collected by the Answer ALS Consortium, these studies were able to rigorously test whether the observed changes might be artifacts related to the techniques of sample collection, storage, processing, and analysis, or truly reflective of important biology,” says Lyle Ostrow, an associate professor of neurology at the Lewis Katz School of Medicine at Temple University, who was not involved in the study. “They developed standard ways to control for these variables, to make sure the results can be accurately compared. Such studies are incredibly important for accelerating ALS therapy development, as they will enable data and samples collected from different studies to be analyzed together.”
Targeted drugs
The researchers now hope to further investigate these genomic regions and see how they might drive different aspects of ALS progression in different subsets of patients. This could help scientists develop drugs that might work in different groups of patients, and help them identify which patients should be chosen for clinical trials of those drugs, based on genetic or epigenetic markers.
Last year, the U.S. Food and Drug Administration approved a drug called tofersen, which can be used in ALS patients with a mutation in a gene called SOD1. This drug is very effective for those patients, who make up about 1 percent of the total population of people with ALS. Fraenkel’s hope is that more drugs can be developed for, and tested in, people with other genetic drivers of ALS.
“If you had a drug like tofersen that works for 1 percent of patients and you just gave it to a typical phase two clinical trial, you probably wouldn’t have anybody with that mutation in the trial, and it would’ve failed. And so that drug, which is a lifesaver for people, would never have gotten through,” Fraenkel says.
The MIT team is now using an approach called quantitative trait locus (QTL) analysis to try to identify subgroups of ALS patients whose disease is driven by specific genomic variants.
“We can integrate the genomics, the transcriptomics, and the epigenomics, as a way to find subgroups of ALS patients who have distinct phenotypic signatures from other ALS patients and healthy controls,” Tsitkov says. “We have already found a few potential hits in that direction.”
The research was funded by the Answer ALS program, which is supported by the Robert Packard Center for ALS Research at Johns Hopkins University, Travelers Insurance, ALS Finding a Cure Foundation, Stay Strong Vs. ALS, Answer ALS Foundation, Microsoft, Caterpillar Foundation, American Airlines, Team Gleason, the U.S. National Institutes of Health, Fishman Family Foundation, Aviators Against ALS, AbbVie Foundation, Chan Zuckerberg Initiative, ALS Association, National Football League, F. Prime, M. Armstrong, Bruce Edwards Foundation, the Judith and Jean Pape Adams Charitable Foundation, Muscular Dystrophy Association, Les Turner ALS Foundation, PGA Tour, Gates Ventures, and Bari Lipp Foundation. This work was also supported, in part, by grants from the National Institutes of Health and the MIT-GSK Gertrude B. Elion Research Fellowship Program for Drug Discovery and Disease.
0 notes
Text
Epigenomic analysis sheds light on risk factors for ALS
New Post has been published on https://thedigitalinsider.com/epigenomic-analysis-sheds-light-on-risk-factors-for-als/
Epigenomic analysis sheds light on risk factors for ALS
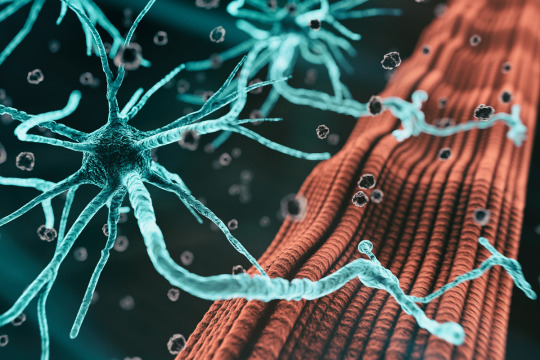

For most patients, it’s unknown exactly what causes amyotrophic lateral sclerosis (ALS), a disease characterized by degeneration of motor neurons that impairs muscle control and eventually leads to death.
Studies have identified certain genes that confer a higher risk of the disease, but scientists believe there are many more genetic risk factors that have yet to be discovered. One reason why these drivers have been hard to find is that some are found in very few patients, making it hard to pick them out without a very large sample of patients. Additionally, some of the risk may be driven by epigenomic factors, rather than mutations in protein-coding genes.
Working with the Answer ALS consortium, a team of MIT researchers has analyzed epigenetic modifications — tags that determine which genes are turned on in a cell — in motor neurons derived from induced pluripotent stem (IPS) cells from 380 ALS patients.
This analysis revealed a strong differential signal associated with a known subtype of ALS, and about 30 locations with modifications that appear to be linked to rates of disease progression in ALS patients. The findings may help scientists develop new treatments that are targeted to patients with certain genetic risk factors.
“If the root causes are different for all these different versions of the disease, the drugs will be very different and the signals in IPS cells will be very different,” says Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering and the senior author of the study. “We may get to a point in a decade or so where we don’t even think of ALS as one disease, where there are drugs that are treating specific types of ALS that only work for one group of patients and not for another.”
MIT postdoc Stanislav Tsitkov is the lead author of the paper, which appears today in Nature Communications.
Finding risk factors
ALS is a rare disease that is estimated to affect about 30,000 people in the United States. One of the challenges in studying the disease is that while genetic variants are believed to account for about 50 percent of ALS risk (with environmental factors making up the rest), most of the variants that contribute to that risk have not been identified.
Similar to Alzheimer’s disease, there may be a large number of genetic variants that can confer risk, but each individual patient may carry only a small number of those. This makes it difficult to identify the risk factors unless scientists have a very large population of patients to analyze.
“Because we expect the disease to be heterogeneous, you need to have large numbers of patients before you can pick up on signals like this. To really be able to classify the subtypes of disease, we’re going to need to look at a lot of people,” Fraenkel says.
About 10 years ago, the Answer ALS consortium began to collect large numbers of patient samples, which could allow for larger-scale studies that might reveal some of the genetic drivers of the disease. From blood samples, researchers can create induced pluripotent stem cells and then induce them to differentiate into motor neurons, the cells most affected by ALS.
“We don’t think all ALS patients are going to be the same, just like all cancers are not the same. And the goal is being able to find drivers of the disease that could be therapeutic targets,” Fraenkel says.
In this study, Fraenkel and his colleagues wanted to see if patient-derived cells could offer any information about molecular differences that are relevant to ALS. They focused on epigenomic modifications, using a method called ATAC-seq to measure chromatin density across the genome of each cell. Chromatin is a complex of DNA and proteins that determines which genes are accessible to be transcribed by the cell, depending on how densely packed the chromatin is.
In data that were collected and analyzed over several years, the researchers did not find any global signal that clearly differentiated the 380 ALS patients in their study from 80 healthy control subjects. However, they did find a strong differential signal associated with a subtype of ALS, characterized by a genetic mutation in the C9orf72 gene.
Additionally, they identified about 30 regions that were associated with slower rates of disease progression in ALS patients. Many of these regions are located near genes related to the cellular inflammatory response; interestingly, several of the identified genes have also been implicated in other neurodegenerative diseases, such as Parkinson’s disease.
“You can use a small number of these epigenomic regions and look at the intensity of the signal there, and predict how quickly someone’s disease will progress. That really validates the hypothesis that the epigenomics can be used as a filter to better understand the contribution of the person’s genome,” Fraenkel says.
“By harnessing the very large number of participant samples and extensive data collected by the Answer ALS Consortium, these studies were able to rigorously test whether the observed changes might be artifacts related to the techniques of sample collection, storage, processing, and analysis, or truly reflective of important biology,” says Lyle Ostrow, an associate professor of neurology at the Lewis Katz School of Medicine at Temple University, who was not involved in the study. “They developed standard ways to control for these variables, to make sure the results can be accurately compared. Such studies are incredibly important for accelerating ALS therapy development, as they will enable data and samples collected from different studies to be analyzed together.”
Targeted drugs
The researchers now hope to further investigate these genomic regions and see how they might drive different aspects of ALS progression in different subsets of patients. This could help scientists develop drugs that might work in different groups of patients, and help them identify which patients should be chosen for clinical trials of those drugs, based on genetic or epigenetic markers.
Last year, the U.S. Food and Drug Administration approved a drug called tofersen, which can be used in ALS patients with a mutation in a gene called SOD1. This drug is very effective for those patients, who make up about 1 percent of the total population of people with ALS. Fraenkel’s hope is that more drugs can be developed for, and tested in, people with other genetic drivers of ALS.
“If you had a drug like tofersen that works for 1 percent of patients and you just gave it to a typical phase two clinical trial, you probably wouldn’t have anybody with that mutation in the trial, and it would’ve failed. And so that drug, which is a lifesaver for people, would never have gotten through,” Fraenkel says.
The MIT team is now using an approach called quantitative trait locus (QTL) analysis to try to identify subgroups of ALS patients whose disease is driven by specific genomic variants.
“We can integrate the genomics, the transcriptomics, and the epigenomics, as a way to find subgroups of ALS patients who have distinct phenotypic signatures from other ALS patients and healthy controls,” Tsitkov says. “We have already found a few potential hits in that direction.”
The research was funded by the Answer ALS program, which is supported by the Robert Packard Center for ALS Research at Johns Hopkins University, Travelers Insurance, ALS Finding a Cure Foundation, Stay Strong Vs. ALS, Answer ALS Foundation, Microsoft, Caterpillar Foundation, American Airlines, Team Gleason, the U.S. National Institutes of Health, Fishman Family Foundation, Aviators Against ALS, AbbVie Foundation, Chan Zuckerberg Initiative, ALS Association, National Football League, F. Prime, M. Armstrong, Bruce Edwards Foundation, the Judith and Jean Pape Adams Charitable Foundation, Muscular Dystrophy Association, Les Turner ALS Foundation, PGA Tour, Gates Ventures, and Bari Lipp Foundation. This work was also supported, in part, by grants from the National Institutes of Health and the MIT-GSK Gertrude B. Elion Research Fellowship Program for Drug Discovery and Disease.
#000#Administration#als#American Airlines#amyotrophic lateral sclerosis#Amyotrophic lateral sclerosis (ALS)#Analysis#approach#Biological engineering#Biology#blood#caterpillar#cell#Cells#chemical#chromatin#coding#communications#data#development#direction#Disease#Diseases#DNA#drug#drug discovery#drugs#engineering#Environmental#filter
0 notes
Text
Deciphering the cellular mechanisms behind ALS
At a time in which scientific research is increasingly cross-disciplinary, Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering, stands out as both a very early adopter of drawing from different scientific fields and a great advocate of the practice today.
When Fraenkel’s students find themselves at an impasse in their…
View On WordPress
0 notes
Photo

003 - VIRUS – FICHAS (037-055)
037 - A finales del siglo XIX, los virus se definían en función de su infectividad, su filtrabilidad y su necesidad de huéspedes vivientes.
038 - Los virus solo habían sido cultivados en plantas y animales. En 1906, Ross Granville Harrison inventó un método para cultivar tejidos en linfa, y, en 1913, E. Steinhardt y colaboradores utilizaron este método para cultivar virus Vaccinia en fragmentos de córnea de cobaya.
039 - En 1928, H. B. Maitland y M. C. Maitland cultivaron el mismo virus en suspensiones de riñones picados de gallina. Su método no fue adoptado ampliamente hasta 1950, cuando se empezó a cultivar poliovirus a gran escala para la producción de vacunas.
040 - Otro avance se produjo en 1931, cuando el patólogo estadounidense Ernest William Goodpasture cultivó el virus de la gripe y otros virus en huevos fertilizados de gallina.
041 - En 1949, John Franklin Enders, Thomas Weller y Frederick Robbins, cultivaron virus de la polio en células cultivadas de embriones humanos, y ésta fue la primera vez que se cultivó un virus sin utilizar tejidos animales sólidos o huevos. Este trabajo permitió a Jonas Salk crear una vacuna efectiva contra la polio.
042 - Con la invención de la microscopía electrónica en 1931 por parte de los ingenieros alemanes Ernst Ruska y Max Knoll, se obtuvieron las primeras imágenes de virus.
043 - En 1935, el bioquímico y virólogo estadounidense Wendell Stanley examinó el virus del mosaico del tabaco y descubrió que estaba compuesto principalmente de proteínas.
044 - Poco tiempo después, el virus fue separado en sus partes de proteínas y de ARN. El virus del mosaico del tabaco fue uno de los primeros en ser cristalizados y, por tanto, la primera estructura que pudo ser observada en detalle. Las primeras imágenes por difracción de rayos X del virus cristalizado las obtuvieron Bernal y Fankuchen en 1941.
045 - Basándose en sus imágenes, Rosalind Franklin descubrió la estructura completa del virus en 1955.
046 - El mismo año, Heinz Fraenkel-Conrat y Robley C. Williams demostraron que el ARN purificado del virus del mosaico del tabaco y sus proteínas de envoltura pueden reproducirse por sí solos, formando virus funcionales, y sugirieron que este debía de ser el modo en que los virus se reproducían en las células huéspedes.
047 - La segunda mitad del siglo XX fue la edad dorada del descubrimiento de virus, y la mayoría de las 2000 especies reconocidas de virus animales, vegetales y bacterianos se descubrieron durante estos años.
048 - En 1957, se descubrieron el arterivirus equino y la causa de la diarrea vírica bovina (un pestivirus).
049 - En 1963 Baruch Blumberg, descubrió el virus de la hepatitis B, y en 1965 Howard Temin describió el primer retrovirus.
050 - La transcriptasa inversa, enzima clave que utilizan los retrovirus para convertir su ARN en ADN, fue descrita originalmente en 1970, de manera independiente por Howard Temin y David Baltimore.
051 - En 1983, el equipo de Luc Montagnier del Instituto Pasteur de Francia aisló por primera vez el retrovirus, actualmente llamado VIH.
052 - 0rigen - Al estudiar el origen de los virus, hay que considerar previamente que los virus son agentes infecciosos acelulares que infectan células y producen viriones para difundir sus genes; por lo que en su origen se debe considerar la interacción entre el virus y su huésped.
053 - Igualmente destaca que la mayoría de las proteínas virales no tienen homólogos en las células modernas, en contradicción con la visión tradicional de los virus como los «ladrones de genes celulares».
054 - Esto sugiere que los genes virales básicamente tienen su origen durante la multiplicación de los genomas virales, o provendrían de linajes celulares ahora extintos (ya que algunas proteínas virales específicas están presentes en virus que infectan a los miembros de los tres dominios de la vida, lo que sugiere que los virus son en realidad muy antiguos en la historia de la vida).
055 - En particular, los análisis estructurales de proteínas de la cápside han revelado que al menos dos tipos de viriones se habrían originado de manera independiente antes que LUCA (el Último antepasado común universal de la vida celular). [email protected]
0 notes
Text
Using Machine Learning to Improve Subseasonal Climate Forecasting
Using Machine Learning to Improve Subseasonal Climate Forecasting
Judah Cohen, director of seasonal forecasting at AER (Atmospheric and Environmental Research) and visiting scientist in MIT’s Department of Civil and Environmental Engineering, and Ernest Fraenkel, professor of biological engineering at MIT, have won first place in three out of four temperature forecasting categories in the Sub-Seasonal Climate Forecast Rodeo competition, hosted by the National…
View On WordPress
0 notes
Text
Protein analysis uncovers new medulloblastoma subtypes
Two patients with the same kind of tumor can have very different experiences. One patient’s cancer may progress quickly while the other grows slowly. Treatments may shrink tumors or have no effect at all. And some patients survive while others don’t.
Efforts to profile tumors at the DNA, RNA, or epigenetic levels have revealed subtypes of tumors that help oncologists diagnose and prognose cancer, but it’s often unclear how to turn that molecular knowledge into new therapeutics. The problem is especially acute for cancers with no clear genetic cause like medulloblastoma, a pediatric brain tumor with toxic treatments and unpredictable outcomes. Now a new effort to look beyond the genome and analyze the tumor’s proteins — the functional players of the cell — has revealed previously unrecognized subtypes of medulloblastoma that could be relevant in the clinic.
Led by scientists at MIT, Boston Children’s Hospital, and the Broad Institute of MIT and Harvard, the study combined clinical, technical, and bioinformatic expertise to show that “multi-omic” approaches aimed at not just genetic material, but also proteins and their modifications, can discover new biomarkers or drug targets.
“We have known since the early days of molecular biology that critical regulatory events take place after genes are transcribed into proteins,” says Ernest Fraenkel, a co-senior author of the new paper, an associate member of the Broad Institute, and professor of biological engineering at MIT. “It’s always been a high priority to look for these events in cancer.”
The researchers describe their findings in the journal Cancer Cell. They have also generated a rich dataset and shared it with the scientific community.
To systematically examine protein modifications in medulloblastoma, the team began by collecting 45 samples of the tumor, with clinical leadership from co-senior author Scott Pomeroy, who is neurologist-in-chief at Boston Children’s Hospital and the Bronson Crothers Professor of Neurology at Harvard Medical School, and co-first author Tenley Archer, a postdoctoral researcher at Children’s Hospital.
The samples were sent to a team in the Broad Institute Proteomics Platform, led by platform senior director and co-senior author Steven Carr. With leadership from co-first author Filip Mundt, the team used a workflow developed at the Broad to perform global measurement of the tumors’ proteins, as well as several ways in which proteins can be modified in the cell after they’re made, such as phosphorylation and acetylation. This proteomics data was then integrated with global measurements of the DNA and RNA by Broad proteomics computational scientists D.R. Mani and Karsten Krug in collaboration with the German Cancer Research Center.
“Recent advances in proteomic technology, especially with respect to analyses of modifications like phosphorylation, enabled us to conduct one of the most comprehensive proteomic studies of medulloblastoma to date, giving us a deeper view of this disease than we could get using genomic methods alone,” says Carr.
Although only a few dozen samples were tested, the results include hundreds of thousands of diverse measurements. This complex dataset was analyzed by a team of computational biologists led by co-senior author Jill Mesirov, a Broad senior institute fellow and professor of medicine at the University of California at San Diego School of Medicine, and Ernest Fraenkel.
Within the four existing subgroups of medulloblastoma — wingless (WNT), sonic hedgehog (SHH), group 3, and group 4 — the researchers found new subtypes.
“Adding data on proteins and their modifications allowed us to see differences in medulloblastoma tumor sub-groups that we can’t see through RNA analyses,” says Pomeroy.
The data showed that group 3 tumors can be divided into two subtypes, including one that is driven by mutations related to the MYC oncogene and has very poor prognosis.
“Notably, the proteomic data improved our ability to stratify the group-3, poorest outcome patients, compared to our previous work using gene expression data,” says Mesirov.
Proteomic analysis of SHH tumors showed different sets of activated pathways. One subtype was strongly driven by the sonic hedgehog pathway, and the other not as much.
To help interpret the data, Fraenkel and co-first author Tobias Ehrenberger led development of a new approach to predict key regulatory proteins that caused changes seen in the proteomic data. Dubbed “kinome analysis,” the methods led them to discover a protein, PRKDC, that when inactivated made one subtype of the tumors more sensitive to radiation treatment.
“Kinome analysis suggested potential therapeutic avenues for type 3 medulloblastoma, a subtype that is known to have poor outcomes — namely, a target that may sensitize tumor cells to radiation,” Pomeroy says.
Fraenkel says patients with medulloblastoma “need better treatments.”
“We hope that insights from this study may lead to therapeutic strategies that result in fewer long-term side effects,” he says.
Protein analysis uncovers new medulloblastoma subtypes syndicated from https://osmowaterfilters.blogspot.com/
0 notes
Text
Books On Books Collection - West Court Gallery, Jesus College, Cambridge UK, 21 May - 27 June 2022
Books On Books Collection – West Court Gallery, Jesus College, Cambridge UK, 21 May – 27 June 2022
Jessica Berenbeim, a University Lecturer at the Faculty of English and a Fellow of Jesus College, has selected works from the Books On Books Collection for this exhibition. With the assistance of Justine Provino, a doctoral student at Oxford and Cambridge, Berenbeim has arranged the works to effect a certain conversation. As she writes,
Artists’ experiments with books and letters have taken many…

View On WordPress
#André Masson#Ernest Fraenkel#Estelle J.#Honorine Tepfer#Isabella Checcaglini#Jérémie Bennequin#Jessica Berenbeim#Justine Provino#Man Ray#Michalis Pichler#Michel Lorand#Mitsou Ronat#Mohammed Bennis#Reinhold Nasshan#Robert Filliou#Rodney Graham#Tibor Papp
0 notes
Text
Deciphering the cellular mechanisms behind ALS
New Post has been published on https://thedigitalinsider.com/deciphering-the-cellular-mechanisms-behind-als/
Deciphering the cellular mechanisms behind ALS


At a time in which scientific research is increasingly cross-disciplinary, Ernest Fraenkel, the Grover M. Hermann Professor in Health Sciences and Technology in MIT’s Department of Biological Engineering, stands out as both a very early adopter of drawing from different scientific fields and a great advocate of the practice today.
When Fraenkel’s students find themselves at an impasse in their work, he suggests they approach their problem from a different angle or look for inspiration in a completely unrelated field.
“I think the thing that I always come back to is try going around it from the side,” Fraenkel says. “Everyone in the field is working in exactly the same way. Maybe you’ll come up with a solution by doing something different.”
Fraenkel’s work untangling the often-complicated mechanisms of disease to develop targeted therapies employs methods from the world of computer science, including algorithms that bring focus to processes most likely to be relevant. Using such methods, he has decoded fundamental aspects of Huntington’s disease and glioblastoma, and he and his collaborators are working to understand the mechanisms behind amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease.
Very early on, Fraenkel was exposed to a merging of scientific disciplines. One of his teachers in high school, who was a student at Columbia University, started a program in which chemistry, physics, and biology were taught together. The teacher encouraged Fraenkel to visit a lab at Columbia run by Cyrus Levinthal, a physicist who taught one of the first biophysics classes at MIT. Fraenkel not only worked at the lab for a summer, he left high school (later earning an equivalency diploma) and started working at the lab full time and taking classes at Columbia.
“Here was a lab that was studying really important questions in biology, but the head of it had trained in physics,” Fraenkel says. “The idea that you could get really important insights by cross-fertilization, that’s something that I’ve always really appreciated. And now, we can see how this approach can impact how people are being treated for diseases or reveal really important fundamentals of science.”
Breaking barriers
At MIT, Fraenkel works in the Department of Biological Engineering and co-directs the Computational Systems Biology graduate program. For the study of ALS, he and his collaborators at Massachusetts General Hospital (MGH), including neurologist and neuroscientist Merit Cudkowicz, were recently awarded $1.25 million each from the nonprofit EverythingALS organization. The strategy behind the gift, Fraenkel says, is to encourage MIT and MGH to increase their collaboration, eventually enlisting other organizations as well, to form a hub for ALS research “to break down barriers in the field and really focus on the core problems.”
Fraenkel has been working with EverythingALS and their data scientists in collaboration with doctors James Berry of MGH and Lyle Ostrow of Temple University. He also works extensively with the nonprofit Answer ALS, a consortium of scientists studying the disease.
Fraenkel first got interested in ALS and other neurodegenerative diseases because traditional molecular biology research had not yielded effective therapies or, in the case of ALS, much insight into the disease’s causes.
“I was interested in places where the traditional approaches of molecular biology” — in which researchers hypothesize that a certain protein or gene or pathway is key to understanding a disease — “were not having a lot of luck or impact,” Fraenkel says. “Those are the places where if you come at it from another direction, the field could really advance.”
Fraenkel says that while traditional molecular biology has produced many valuable discoveries, it’s not very systematic. “If you start with the wrong hypothesis, you’re not going to get very far,” he says.
Systems biology, on the other hand, measures many cellular changes — including transcription of genes, protein-DNA interactions, of thousands of chemical compounds and of protein modifications — and can apply artificial intelligence and machine learning to those measurements to collectively identify the most important interactions.
“The goal of systems biology is to systematically measure as many cellular changes as possible, integrate this data, and let the data guide you to the most promising hypotheses,” Fraenkel says.
The Answer ALS project, with which Frankel works, involves approximately a thousand people with ALS who provided clinical information about their disease and blood cells. Their blood cells were reprogrammed to be pluripotent stem cells, meaning that the cells could be used to grow neurons that are studied and compared to neurons from a control group.
Emotional connection
While Fraenkel was intellectually inspired to apply systems biology to the challenging problem of understanding ALS — there is no known cause or cure for 80 to 90 percent of people with ALS — he also felt a strong emotional connection to the community of people with ALS and their advocates.
He tells a story of going to meet the director of an ALS organization in Israel who was trying to encourage scientists to work on the disease. Fraenkel knew the man had ALS. What he didn’t know before arriving at the meeting was that he was immobilized, lying in a hospital bed in his living room and only able to communicate with eye-blinking software.
“I sat down so we could both see the screen he was using to type characters out,” Fraenkel says, “and we had this fascinating conversation.”
“Here was a young guy in the prime of life, suffering in a way that’s unimaginable. At the same time, he was doing something amazing, running this organization to try to make a change. And he wasn’t the only one,” he says. “You meet one, and then another and then another — people who are sometimes on their last breaths and are still pushing to make a difference and cure the disease.”
The gift from EverythingALS — which was founded by Indu Navar after losing her husband, Peter Cohen, to ALS and later merged with CureALS, founded by Bill Nuti, who is living with ALS — aims to research the root causes of the disease, in the hope of finding therapies to stop its progression, and natural healing processes that could possibly restore function of damaged nerves.
To achieve those goals, Fraenkel says it is crucial to measure molecular changes in the cells of people with ALS and also to quantify the symptoms of ALS, which presents very differently from person to person. Fraenkel refers to how understanding the differences in various types of cancer has led to much better treatments, pointing out that ALS is nowhere near as well categorized or understood.
“The subtyping is really going to be what the field needs,” he says. “The prognosis for more than 80 percent of people with ALS is not appreciably different than it would have been 20, or maybe even 100, years ago.”
In the same way that Fraenkel was fascinated as a high school student by doing biology in a physicist’s lab, he says he loves that at MIT, different disciplines work together easily.
“You reach out to MIT colleagues in other departments, and they’re not surprised to hear from someone who’s not in their field,” Fraenkel says. “We’re a goal-oriented institution that focuses on solving hard problems.”
#Algorithms#als#amazing#amyotrophic lateral sclerosis#Amyotrophic lateral sclerosis (ALS)#approach#artificial#Artificial Intelligence#Biological engineering#Biology#biophysics#blood#breaths#Cancer#Cells#change#chemical#chemical compounds#chemistry#classes#Collaboration#Community#computation#computer#Computer Science#Computer science and technology#data#direction#Discoveries#Disease
0 notes
Text
Books On Books Collection - Michel Lorand
Books On Books Collection – Michel Lorand
Après Un Coup de Dés (2015)
Après Un Coup de Dés (2015)Michel LorandCover and gatherings, untrimmed and unbound, in glassine envelope. Cover: H362 x W260; gatherings: H362 x W256 mm; 32 unnumbered pages. Edition of 50, of which this is #19. Acquired from the artist, 22 October 2021.Photos: Books On Books Collection. Displayed with the artist’s permission.
Since the 1960s when Ernest Fraenkel,…

View On WordPress
#André Masson#Aurélie Noury#Benjamin Lord#Brian Larosche#Cerith Wyn Evans#Chris Edwards#Christopher Brennan#David Dernie#Derek Beaulieu#Eric Zboya#Ernest Fraenkel#Filiep Tacq#Gottfried Benn#Guido Molinari#Ian Tyson & Neil Crawford#Jacques Sojcher#Jacques Vernière#Jérémie Bennequin#Jeff Clark & Robert Bononno#Jim Clinefelter#John Keats#Kathy Bruce#Marcel Broodthaers#Mario Diacono#Martin Heidegger#Michael Maranda#Michalis Pichler#Michel Lorand#Mitsou Ronat#Nicolas Guyot
5 notes
·
View notes
Text
Books On Books Collection - Rémi Forte
Books On Books Collection – Rémi Forte
The Spectre of Mallarmé (2022)
The Spectre of Mallarmé (2022)Rémi FortePaper booklet, sewn. H190 x W126 mm, 16 pages. Edition of 50. Acquired from Rémi Forte, 8 April 2022.Photos: Books On Books Collection. Displayed with permission of the artist.
Published by Derek Beaulieu’s No Press in 2022, Rémi Forte’s images actually began life digitally over a ten-year period, first emerging on Instagram…

View On WordPress
0 notes
Text
Bookmarking Book Art - Mario Diacono
Bookmarking Book Art – Mario Diacono
a METRICA n’aboolira (1968)
a METRICA n’aboolira (1968)Mario DiaconoPapercover, stapled. H x W mm, unnumbered pages.Photos: Arengario, displayed with permission of the artist.
Besides being first out of the gate with an “homage by redaction” of Un Coup de Dés, Mario Diacono is perhaps the first hommageur to give a sociopolitical cast to the effort. In an interview in Ursula, Diacono…

View On WordPress
#Danièle Huillet#Didier Mutel#Ernest Fraenkel#Jean-Marie Straub#Marcel Broodthaers#Mario Diacono#Stéphane Mallarmé
0 notes
Text
Books On Books Collection - Ernest Fraenkel
Books On Books Collection – Ernest Fraenkel
Les Dessins Trans-conscients de Stéphane Mallarméà propos de la Typographie de Un Coup de Dés (1960)
Les Dessins Trans-conscients de Stéphane Mallarmé, à propos de la Typographie de Un Coup de Dés (1960)Ernest FraenkelPaperback, stapled to fold-in sleeve. H245 x W160 mm, 44 pages bound with 68 pages on 8.5 uncut folded and gathered sheets. Acquired from À la Page, 12 October 2021.Photos: Books…

View On WordPress
#Andy Simionato#Cosmopolis#David W. Seaman#Ernest Fraenkel#Geraldo de Barros#Giulio Maffei#Jed Rasula#Jorge Méndez Blake#Karen Ann Donnachie#Kathy Bruce#Man Ray#Marcel Broodthaers#Mario Diacono#Stéphane Mallarmé#Tayyib Yavuz
0 notes
Text
Books On Books Collection - Mitsou Ronat & Tibor Papp
Books On Books Collection – Mitsou Ronat & Tibor Papp
Un Coup de Dés Jamais N’abolira le Hasard (1980)

Poème: Un coup de Dés jamais n’abolira le Hasard par Stéphane Mallarmé (1980)
Édition Mise en Oeuvre et Présentée par Mitsou Ronat, Réalisée par Tibor Papp.
Two sets of folded & gathered folios, enclosed in a portfolio with four flaps; Portfolio: H380 x W285 mm; Folios: H380 x W285 mm; Poème, 24 pages, including the cover; “Le Genre …”, 28 pages,…
View On WordPress
#Ambroise Vollard#Aurélie Noury#Bruno Montels#Claude Minière#Ernest Fraenkel#Françoise Morel#Jacques Roubaud#Jean Pierre Faye#Le Groupe d&039;atelier#Man Ray#Michel Pierson#Mitsou Ronat#Paul Nagy#Philippe Dôme#Rodolfo Hinostroza#Stéphane Mallarmé#Sylvain Moore#Tibor Papp
0 notes
Text
Books On Books Collection - Eric Zboya
Books On Books Collection – Eric Zboya
un coup de dés jamais n’abolira le hasard: translations (2018)

un coup de dés jamais n’abolira le hasard: translations (2018)
Eric Zboya
Perfect bound; 60 pages printed on Rives Design 170 gsm Brilliant White.
H280 x W204 x D85 mm. Edition of 120 of which this is #42 and signed. Acquired from the artist, 1 April 2019.

Eric Zboya is poet, writer and artist. Years before this book, he wrote for
View On WordPress
#Andrew Michael Roberts#Anna Katharine Schaffner#C. T. Funkhouser#Charles Baudelaire#Dick Higgins#Didier Mutel#Eric Zboya#Ernest Fraenkel#Guido Molinari#Jed Rasula#Kim Knowles#Marcel Broodthaers#Michalis Pichler#Paul Valéry#Sammy Engramer#Ulrich Weger
0 notes
Text
Books On Book Collection - Sammy Engramer
Books On Book Collection – Sammy Engramer
Un Coup de Dés Jamais N’abolira le Hasard: Wave (2009)
Mallarmé’s strange poem, first published in the London-based journal Cosmopolis in 1897, had to wait until 1914 before appearing in a format close to the one Stéphane Mallarmé envisioned with the gallerist and publisher Ambroise Vollard.





Taking the poem’s self-referential line about its words appearing as a constellation, first Ernest…
View On WordPress
#Ambroise Vollard#Andrew Michael Roberts#Anna Katharine Schaffner#Dick Higgins#Eric Zboya#Ernest Fraenkel#Jed Rasula#Kim Knowles#Marcel Broodthaers#Mario Diacono#Michalis Pichler#Sam Halliday#Sammy Engramer#Stéphane Mallarmé#Ulrich Weger
0 notes