#Non-Coding RNAs
Explore tagged Tumblr posts
Text
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra #GeneExpression #BiologyClass #GenomicImprinting #NonCodingRNAs #CollaborativeLearning
Dr. Mishra: “Hello there! I see we have a new face in our biology class today. Welcome! We’re so glad you’re here, and I want you to know that this classroom is a friendly and supportive place. If you ever have questions or need assistance, don’t hesitate to ask me or your classmates. We’re all here to learn together and make this an enjoyable experience for you.” Abby: “Hello. My name is Abha…

View On WordPress
#biology#Chromatin Structure#Collaborative Learning#Epigenetic Modifications#Gene expression#Genomic Imprinting#MicroRNAs#Non-Coding RNAs#Transcription Factors
0 notes
Text
youtube
#Osteoclasts#osteoblasts#epigenetic regulation#long non-coding RNAs#lncRNAs#bone remodeling#titanium implants#nanotopography#bone regeneration#implant integration#bone healing#osteoimmunology#epigenetics#cellular interactions#biomaterials#tissue engineering#orthopedic research#regenerative medicine#gene expression#implant materials.#Youtube
0 notes
Text
Τhe RΝΑ World
Every now and again, something turns up that makes one feel good. In my most recent case it was a book review. No, not one of my books, but one in Nature (vol 632, pp 250-251) on RNA. The Nobel prize winner, Thomas Cech describes RNA as “Folding into origami-like shapes, it can pull off wild stunts that make its genetic parent, DNA, look like a one-trick pony.” This is in his book The Catalyst…
0 notes
Text
Giant pandas have digestive systems that are typical for carnivores. Yet, bamboo is their main source of food. They have evolved several features; for example, pseudo thumbs to grasp bamboo and flat teeth that are well suited for crushing it, that make it possible for them to live off plants. All living organisms have DNA, which stores the genetic information in a cell, and RNA, which carries and transfers this information. MicroRNAs (miRNA) are small non-coding RNAs that play an important role in gene expression, the process of turning the information encoded in a gene into a function. MiRNA from plants can be absorbed through food.
Continue Reading.
45 notes
·
View notes
Text

Epigenetics: A Journey Through Inheritance Beyond Genes
For centuries, scientists have been fascinated by the mysteries of heredity and how traits are passed down from generation to generation. DNA, the molecule that stores our genetic code, was once thought to be the sole determinant of our characteristics. However, a new frontier in biology, revealing a captivating layer of complexity beyond the DNA sequence itself: Epigenetics.
What is Epigenetics?
The term "epigenetics" was first coined in the 1940s by British biologist Conrad Waddington, but it wasn't until the late 20th century that its significance truly blossomed. Epigenetics, literally meaning "above genetics," refers to the study of heritable changes in gene expression that occur without alterations to the DNA sequence itself. Imagine DNA as the musical score, but epigenetics are the conductor and musicians who determine how the music is played. Through chemical modifications and adjustments to the proteins around DNA, epigenetics dictates which genes are turned on or off, influencing how cells function and ultimately shaping our health, development, and even behavior. Think of your DNA as the hardware: it contains the basic instructions for building and running your body. But epigenetics acts like the software, fine-tuning those instructions and determining which genes get turned on or off at specific times and in specific cells. These modifications, like chemical tags or changes in the packaging of DNA, don't alter the underlying code itself, but they can have a profound impact on how it's read and interpreted.
The Key Players:
DNA methylation: This process involves adding a methyl group to DNA, essentially silencing the gene it's attached to. Imagine it like putting a dimmer switch on a light bulb.
Histone modifications: Histones are proteins that package DNA, and changes in their structure can make genes more or less accessible to the cellular machinery needed for expression. Think of it like adjusting the curtains around a window - open wide for full light, slightly closed for filtered light.
Non-coding RNAs: These are molecules that don't code for proteins but can regulate gene expression in various ways. They're like the backstage crew in a play, ensuring everything runs smoothly.
The Power of Epigenetic Regulation
Epigenetic regulation plays a crucial role in various biological processes, including:
Development: During embryonic development, different cell types emerge from the same DNA blueprint by activating or silencing specific gene sets through epigenetic modifications.
Cellular differentiation: Specialized cells like muscle or nerve cells have unique functions due to differences in their active genes, controlled by epigenetic mechanisms.
Learning and memory: Epigenetic changes in brain cells are thought to be essential for learning and forming memories.
Aging: As we age, our epigenome accumulates changes that can contribute to age-related decline and disease.
Environmental influences: Diet, exercise, stress, and exposure to toxins can leave epigenetic marks on our genes, potentially impacting our health and even the health of future generations.
Epigenetics reminds us that we are not simply products of our genes. Our environment, choices, and experiences leave their mark, shaping who we are and potentially influencing our children's health. This deeper understanding of ourselves opens doors for self-awareness, empowerment, and potentially reshaping our narratives – not just as individuals, but as a species with the potential to leave a healthier legacy for generations to come.
#life science#biology#science sculpt#molecular biology#biotechnology#epigenetics#daily dose of science#dna#genetic inheritance#genetics#decoding dna#genetic code#science#double helix
128 notes
·
View notes
Text
Genetic engineering: CRISPR and beyond
In genetic engineering, we find ourselves amidst a scientific revolution with the advent of revolutionary technologies like CRISPR-Cas9. However, our journey into the intricate landscape of genetic manipulation is far from complete. This post delves into the nuanced world of genetic engineering, exploring cutting-edge technologies and their remarkable potential in shaping the future of medicine and biotechnology.
CRISPR-Cas9: Precision at the Molecular Level
CRISPR-Cas9, a revolutionary genome editing tool, stands for Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein 9. It utilizes a guide RNA (gRNA) to target specific DNA sequences, and the Cas9 protein acts as molecular scissors to cut the DNA at precisely defined locations. This break in the DNA prompts the cell's natural repair machinery to make changes, either through non-homologous end joining (NHEJ) or homology-directed repair (HDR). CRISPR-Cas9's precision allows for gene knockout, modification, or insertion with remarkable accuracy.
Beyond CRISPR: Emerging Technologies
While CRISPR-Cas9 has dominated the field of genetic engineering, numerous promising technologies have emerged on the horizon. These include CRISPR-Cas variants like CRISPR-Cas12 and CRISPR-Cas13, which offer unique advantages such as smaller size, increased specificity, and targeting of RNA. Additionally, base editing techniques, such as adenine base editors (ABEs) and cytosine base editors (CBEs), enable the direct conversion of one DNA base into another without causing double-strand breaks, expanding the range of genetic modifications possible.
Applications in Medicine
The implications of these advancements are profound, particularly in medicine. Genetic engineering can potentially treat various genetic disorders, from cystic fibrosis to sickle cell anemia, by correcting disease-causing mutations at their source. Precision medicine, tailored to an individual's genetic makeup, is becoming increasingly feasible, allowing for personalized therapies with minimal side effects.
Ethical Considerations and Regulation
As we venture further into the genetic frontier, we must acknowledge the ethical considerations surrounding genetic engineering. The ability to modify the human germline, with implications for future generations, raises ethical dilemmas that necessitate rigorous oversight and regulation. The international community is developing guidelines to ensure responsible use of these powerful tools.
Future Directions and Challenges
While genetic engineering offers immense promise, it is not without its challenges. Off-target effects, unintended consequences, and the potential for creating designer babies are among the issues that demand careful consideration. Researchers and ethicists must work in tandem to navigate this uncharted territory.
References
Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., … & Liu, D. R. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576(7785), 149-157.
Kime, E. (2021). CRISPR and the ethics of gene editing. Nature Reviews Genetics, 22(1), 3-4.
This post only scratches the surface of the profound transformations occurring in genetic engineering. The relentless pursuit of knowledge and ethical exploration will shape the future of this field as we continue to unlock the intricate secrets of our genetic code.

#science#biology#college#education#school#student#medicine#doctors#health#healthcare#genetics#genetic engineering#crispr#ethical genetics
77 notes
·
View notes
Text
Propaganda!
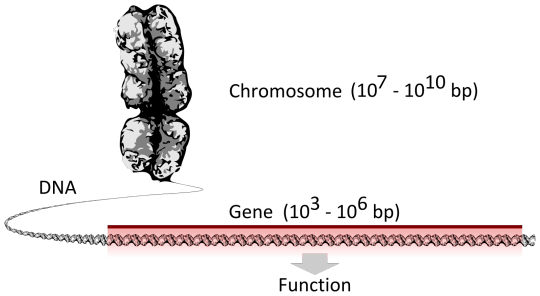

In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression (the synthesis of RNA or protein from a gene), DNA is first copied into RNA. RNA can be directly functional or be the intermediate template for the synthesis of a protein.
Adult stem cells are undifferentiated cells, found throughout the body after development, that multiply by cell division to replenish dying cells and regenerate damaged tissues. Also known as somatic stem cells (from Greek σωματικóς, meaning of the body), they can be found in juvenile, adult animals, and humans, unlike embryonic stem cells.
#genes#Unipotent stem cells#tournament poll#polls#wikipedia#cells of the human body#science tournament#biochemistry
10 notes
·
View notes
Text
Reading a reddit post from someone who alleges they sequenced aliens and some of it is "Ooh this person doesn't understand biology, they talked like there's no such things non-coding RNA" but then I remember that some actual biologists will forget that non-coding RNA exists too.
25 notes
·
View notes
Text
In reference to this post as promised I'm elaborating
Alive is defined by the Oxford Dictionary as;
(of a person, animal, or plant) living, not dead Alert and Active; animated
Merriam-webster defines Alive as;
Having life: not dead or inanimate
Both of these fit what we see with characters such as Boothill, but in biological science, there is more complexity regarding what is or can be defined as alive.
Throughout my science degree, I must consistently refer to the expanded definition. Because of that, I have struggled to define certain organisms in a strictly 'alive' or 'dead' definition. The definition of life is complex and multifaceted and is typically defined by 7 key characteristics, these are
Cellular Organization: All living organisms are composed of one or more cells, which are considered the basic units of life.
Metabolism: Living organisms undergo chemical reactions that allow them to transform energy and materials from their environment into usable forms. This includes processes such as respiration, digestion, and photosynthesis.
Growth and Development: Living organisms grow and develop according to specific instructions coded for by their DNA or RNA.
Reproduction: Living organisms have the ability to reproduce, either sexually or asexually, to produce new individuals of the same species.
Response to Stimuli: Living organisms can respond to environmental stimuli, such as light, temperature, and touch.
Homeostasis: Living organisms maintain a stable internal environment, regulating factors such as temperature, pH, and hydration to sustain life.
Adaptation and Evolution: Living organisms have the ability to adapt to their environment through the process of evolution by natural selection over generations.
These criteria form the basis of our understanding of life. However, some entities like viruses challenge these criteria, as they exhibit some but not all characteristics of life, such as having genetic material and evolving, but lacking cellular structure and metabolism on their own.
Now back on topic, Boothill is in a way pseudo-dead/alive. Shrodiger's cowboy if you will. He exists in the grey area between the definition of living and non-living. Now if we work through the list above and work out the rough percentage of the scientific definition our favourite galaxy ranger fits into. I'll start from the top, as far as we are aware (please correct me if I'm wrong) Boothill is lacking a majority of his original cellular makeup, excluding what's left visible of his skin which I assume isn't synthetic. Assuming all of this he fits the first characteristic poorly and therefore I'm giving him an F+ for effort. The second characteristic is metabolism... I'm going to just instantly fail him because I don't think bullets truly have any nutritional value and therefore he gains no energy from them (I also highly doubt he needs more than a phone charger cable to stop himself from powering down with the Windows shutdown noise). Growth and Development, unless he's made of a biological metal that can grow he also fails in this department, he also lacks a majority of his original biological makeup which also means he fails.
(Boothill isn't doing so well so far...)
Reproduction... I think this one is pretty self-explanatory. I highly doubt there was any need for there to be any reproductive organ and therefore he has no way to reproduce. (Sorry to all the fanfic authors out there that have just had a field day coming up with as many different ways this man can get some, keep being as delusional as you wish I won't judge). Finally, we reach one Boothill can do (YAY!!!) response to stimuli. despite him not having any or very little feeling in his mechanical body parts there is still his face and very pinchable cheeks. I'm also going to treat his lack of feeling as being similar to people with severe nerve damage, still alive and just can't feel shit. Now, fanfics also enjoy this one, homeostasis, it makes sense that he would be able to regulate internal temperature especially because overheating would fry him like a chip in oil. So I'm fully on board with this man being able to sound like my laptop when I use it for too long or open a million different AO3 tabs. (plus a flustered Boothill sounding like an overheating laptop is a hilarious thought to me). the last characteristic is adaption and evolution, this to me links to reproduction as it typically refers to the entire 'species' not just an individual. another fail.
Out of the seven characteristics Boothill fits at best two. This means he's at best 28.5% alive, not a good mark if it were a test but interesting. As far as I'm concerned he's similar to a virus in the sense that despite not fitting into the characteristics that well he's still very much alive.
Apologies for the long read, I had a brainworm and needed to free it.
5 notes
·
View notes
Text
Abstract
Therapeutic applications of synthetic mRNA were proposed more than 30 years ago, and are currently the basis of one of the vaccine platforms used at a massive scale as part of the public health strategy to get COVID-19 under control. To date, there are no published studies on the biodistribution, cellular uptake, endosomal escape, translation rates, functional half-life and inactivation kinetics of synthetic mRNA, rates and duration of vaccine-induced antigen expression in different cell types. Furthermore, despite the assumption that there is no possibility of genomic integration of therapeutic synthetic mRNA, only one recent study has examined interactions between vaccine mRNA and the genome of transfected cells, and reported that an endogenous retrotransposon, LINE-1 is unsilenced following mRNA entry to the cell, leading to reverse transcription of full length vaccine mRNA sequences, and nuclear entry. This finding should be a major safety concern, given the possibility of synthetic mRNA-driven epigenetic and genomic modifications arising. We propose that in susceptible individuals, cytosolic clearance of nucleotide modified synthetic (nms-mRNAs) is impeded. Sustained presence of nms-mRNA in the cytoplasm deregulates and activates endogenous transposable elements (TEs), causing some of the mRNA copies to be reverse transcribed. The cytosolic accumulation of the nms-mRNA and the reverse transcribed cDNA molecules activates RNA and DNA sensory pathways. Their concurrent activation initiates a synchronized innate response against non-self nucleic acids, prompting type-I interferon and pro-inflammatory cytokine production which, if unregulated, leads to autoinflammatory and autoimmune conditions, while activated TEs increase the risk of insertional mutagenesis of the reverse transcribed molecules, which can disrupt coding regions, enhance the risk of mutations in tumour suppressor genes, and lead to sustained DNA damage. Susceptible individuals would then expectedly have an increased risk of DNA damage, chronic autoinflammation, autoimmunity and cancer. In light of the current mass administration of nms-mRNA vaccines, it is essential and urgent to fully understand the intracellular cascades initiated by cellular uptake of synthetic mRNA and the consequences of these molecular events.
4 notes
·
View notes
Text
Genes, Vol. 15, Pages 1: #RNA Polymerases IV and V Are Involved in Olive Fruit Development
Transcription is carried out in most eukaryotes by three multimeric complexes (#RNA polymerases I, II and III). However, plants contain two additional #RNA polymerases (IV and V), which have evolved from #RNA polymerase II. #RNA polymerases II, IV and V contain both common and specific subunits that may specialise some of their functions. In this study, we conducted a search for the genes that putatively code for the specific subunits of #RNA polymerases IV and V, as well as those corresponding to #RNA polymerase II in olive trees. Based on the homology with the genes of Arabidopsis thaliana, we identified 13 genes that putatively code for the specific subunits of polymerases IV and V, and 16 genes that code for the corresponding specific subunits of polymerase II in olives. The transcriptomic analysis by #RNA-Seq revealed that the expression of the #RNA polymerases IV and V genes was induced during the initial stages of fruit development. Given that #RNA polymerases IV and V are involved in the transcription of long non-coding #RNAs, we investigated their expression and observed relevant changes in the expression of this type of #RNAs. Particularly, the expression of the intergenic and intronic long non-coding #RNAs tended to increase in the early steps of fruit development, suggesting their potential role in this process. The positive correlation between the expression of #RNA polymerases IV and V subunits and the expression of non-coding #RNAs supports the hypothesis that #RNA polymerases IV and V may play a role in fruit development through the synthesis of this type of #RNAs. https://www.mdpi.com/2073-4425/15/1/1?utm_source=dlvr.it&utm_medium=tumblr
2 notes
·
View notes
Text
youtube
#lncRNA expression#follicular fluid#exosomes#obesity#polycystic ovary syndrome#PCOS#high-throughput sequencing#gene expression profiling#molecular mechanisms#biomarkers#metabolic syndrome#reproductive health#fertility research#ovarian function#non-coding RNA#genomics#endocrine disorders#therapeutic targets#diagnostic tools#personalized medicine#Youtube
0 notes
Text
RNAs, introns and introduction to recombinant DNA.
The genome is a vault of genetic information, but on its own its kind of useless as the information cannot be used in the cell. Utilization of this information requires a coordinated effort from enzymes and proteins, this coordination of chemical reactions is known as genome expression. The initial step of genome expression is known as the transcriptome, a collection of RNA molecules which is made up of the genes active in the cell at the time. (its constantly changing to the environment that the cell is in) the transcriptome is maintained by transcription a simple process in which individual genes are copied and pasted into RNA molecules known as mRNA. The second product within gene expression is known as proteome which you can think of as the cells library of proteins at hand. This gives the cell its unique and individual characteristics that it can express. The proteins in this library known as the proteome are synthesized by the translation of some RNA molecules.
I think it is of utmost importance to start with the most simple and important step…
Transcription:
The first step in transcription is initiation,
Transcription begins with the binding of RNA polymerase 2 to the promoter region of a gene, the most common promotor contains a conserved gene sequence we call the TATA box, but there are many others that exist which are show by the different TF families.
The next step we have the formation of transcription initiation complex, this is when transcription factors such as TATA-binding protein also known as TBP, bind to the promoter, forming a pre-initiation complex. Other general transcription factors join, and RNA polymerase 2 is recruited to form the transcription initiation complex.
Next, we have initiation of transcription where RNA polymerase 2 unwinds the DNA helix at the transcription “start site”. The enzyme catalyses the synthesis of a short RNA primer (which is about 10 nucleotides in length) complementary to the template DNA strand.
The second step is Elongation:
Once the RNA polymerase has synthesized the initial RNA primer, it proceeds to elongate the RNA chain, The RNA polymerase adds ribonucleotide complementary to the template DNA strand. This process is known as RNA chain Elongation.
After this, we have nucleosome remodelling where the DNA in eukaryotic cells is wrapped around histone proteins to form nucleosomes. As transcription proceeds, nucleosomes are temporarily disrupted and then reassembled, allowing RNA polymerase to access the DNA template.
The third step is Termination:
Eukaryotic mRNA molecules undergo post-transcriptional modification, this includes polyadenylation which involves the addition of a poly-A tail to the 3’ end of the RNA. Polyadenylation is then accompanied by cleavage of the RNA precursor at the specific site.
During this process transcription termination signals are recognized by RNA polymerase 2, leading to its dissociation from the DNA template. The termination signal will most likely include the poly-A signal.
The fourth step being RNA processing:
The first step of RNA processing is known as “capping”. Capping the 5’ end of the newly synthesized mRNA is modified with a 7-methylguanosine cap, the whole point of this cap is just to protect the mRNA as it leaves the nucleus and helps in the transportation process.
The second step is splicing, Eukaryotic genes often contain introns (which are kind of gaps or non-coding regions) and exons (which are the important bits, coding regions). In a process called splicing the annoying useless introns are cut and the exons are then pinched together with each other. This is all catalysed by a complex of RNA and proteins known as spliceosomes.
The fifth step being transport and translation:
This is when mRNA is transported out of the nucleus to the cytoplasm where it serves as a template for translation.
The sixth step is regulation:
Gene expression if tightly regulated at the transcriptional level by various elements, including transcriptional factors and chromatin modification, there is also enhancers and silencers that help regulate elements that influence transcription.
We also have post-transcriptional regulation, which is processes such as alternative splicing and RNA stability which play roles in determining the final mRNA products.
It is important to acknowledge that Eukaryotic transcription is more complex than prokaryotic transcription due to the presence of introns, the involvement of multiple RNA polymerases, and the necessity for additional processing steps.
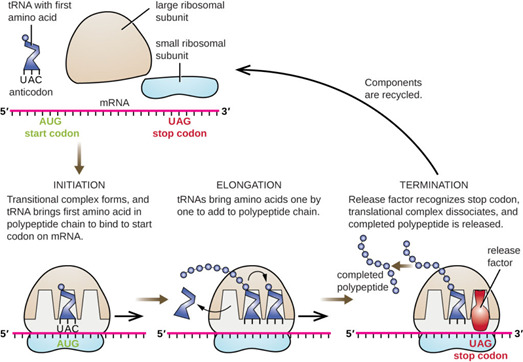
This diagram helps visualise the main steps more simply.
Hope this was clear excuse any mistakes in grammar :)
Referencing:
Picture link above^
2 notes
·
View notes
Text
Bacteriophages suppress CRISPR–Cas immunity using RNA-based anti-CRISPRs
2 notes
·
View notes
Text
RNA: The Dynamic Molecule Driving Life's Diversity
DNA, the blueprint of life, often steals the spotlight when it comes to genetics. But lurking in its shadow is another crucial molecule, RNA (Ribonucleic Acid), playing a pivotal role in the symphony of life. More than just a passive messenger, RNA boasts a vibrant history and holds exciting potential for the future. Let's embark on a journey to unveil the world of RNA, exploring its captivating story and why it deserves your attention.
The story of RNA's discovery began in 1860 when Friedrich Miescher isolated a mysterious "nuclein" from white blood cells. However, it wasn't until the 1950s that James Watson and Francis Crick, alongside Rosalind Franklin (whose contributions were initially overlooked), unraveled the structure of DNA, relegating RNA to a supporting role as a mere messenger molecule. But the plot thickened in the 1960s when researchers like Howard Temin and David Baltimore stumbled upon reverse transcriptase, an enzyme that could convert RNA into DNA, challenging the long-held "central dogma" of DNA being the sole source of genetic information. This discovery opened the door to a whole new understanding of RNA's diverse capabilities.
The Many Faces of RNA
But RNA isn't just a protein puppet master. There are different types of RNA, each with unique jobs:
Messenger RNA (mRNA): Delivers the protein-making message. Transfer RNA (tRNA): Brings the amino acids, the building blocks of proteins, to the party. Ribosomal RNA (rRNA): The foreman of the ribosome factory, making sure everything runs smoothly. Non-coding RNA (ncRNA): A diverse bunch with various roles, from regulating genes to fighting viruses.
The true game-changer came in the early 2000s. Scientists stumbled upon a vast class of non-coding RNAs that don't code for proteins but have diverse and crucial functions. microRNAs (miRNAs), for example, regulate gene expression by silencing specific genes, while long non-coding RNAs (lncRNAs) control various cellular processes like development and disease. This discovery shattered the dogma that only protein-coding genes mattered, highlighting the crucial roles played by non-coding RNAs.
This newfound understanding of RNA's potential has ignited a revolution in medicine. Researchers are exploring RNA-based therapies for various diseases, from cancer and neurodegenerative disorders to viral infections. mRNA vaccines, like the ones used against COVID-19, harness the power of messenger RNA to deliver genetic instructions directly to cells, triggering immune responses. The future holds even more promise, with scientists exploring techniques like CRISPR-Cas9 to edit RNA and potentially treat genetic diseases.
New discoveries are constantly rewriting our understanding of this versatile molecule. Its adaptability and diverse roles make it a powerful tool for exploring the very essence of life, from evolution and development to disease and therapy. So, the next time you hear about genes, remember that RNA, the often-overlooked player, is just as crucial in shaping the tapestry of life. It's a story of constant evolution, unexpected discoveries, and immense potential, making RNA a molecule brimming with fascination and promise for the future.
#science sculpt#life science#science#molecular biology#biotechnology#biology#genetics#RNA#daily dose of science#dailyprompt#meaningful#scientific illustration#the glass scientists#microscopic world#microscopy#artists on tumblr#digital artist
12 notes
·
View notes
Text
interesting! it's not clear from the pull quote, but these vaccines are "RNA-based" in a very different way than the mRNA vaccines we've been using for COVID-19.
mRNA vaccines introduce mRNA sequences that code for a viral protein, causing the body's cells to produce these proteins which then trigger an immune response, most importantly activating B and T immune cells and antibodies, some of which will stick around and be able to specifically recognize that viral protein in the future and attack viruses with that protein. one problem with these vaccines is that, if those viral proteins mutate too much, this protection will be weakened or disappear.
the vaccines discussed in this article, on the other hand, are live viruses altered to get rid of their ability to suppress the body's antiviral RNA interference (RNAi). this is a totally different mechanism of the immune system—and unfortunately my one immunology class only quickly touched on it, so I don't know how it works in great detail. basically, the body's cells recognize that there's viral RNA and produce their own little bits of RNA ("small interfering RNA," or siRNA) that match up with that viral RNA. these are then used in a system that recognizes viral RNA when it enters a cell and stops it from being translated into proteins. I think it's a recent discovery that these antiviral siRNAs continue to circulate in the bloodstream after an infection is cleared. this means that when the mice in this study were vaccinated with viruses altered to be particularly susceptible to RNAi, (1) these viruses didn't harm them because RNAi was able to clear the infection, and (2) the siRNAs that continued to circulate protected them from subsequent infections for at least 90 days.
my (non-expert) takeaways: this is really exciting as a plausible new mechanism for long-lasting immune protection which could also work for people deficient in the B cell/T cell/antibody aspects of the immune system, and in infants! as the article says, it's also more likely to work for different strains of a virus because pieces of siRNA will be produced which match (as far as I can tell) all/most of the viral RNA genome, including sections which aren't likely to mutate. a caveat: this is a proof of concept in mice and with a virus which doesn't actually infect humans. the article notes that humans produce siRNA in reponse to influenza, so it seems likely that this strategy could work against the flu. I don't know the current state of research on RNA interference in humans, so I'm curious whether we know, for example, how long siRNA remains circulating or how effective an RNA interference response is against flu.
the "universal vaccine" statement is a little misleading: a different vaccine would have to be produced for a different virus (but using the same strategy), and I don't think this could work at all against DNA viruses (e.g. chickenpox, herpes, HPV). but definitely a cool approach that I hope works out!
Scientists at UC Riverside have demonstrated a new, RNA-based vaccine strategy that is effective against any strain of a virus and can be used safely even by babies or the immunocompromised. Every year, researchers try to predict the four influenza strains that are most likely to be prevalent during the upcoming flu season. And every year, people line up to get their updated vaccine, hoping the researchers formulated the shot correctly. The same is true of COVID vaccines, which have been reformulated to target sub-variants of the most prevalent strains circulating in the U.S. This new strategy would eliminate the need to create all these different shots, because it targets a part of the viral genome that is common to all strains of a virus. The vaccine, how it works, and a demonstration of its efficacy in mice is described in a paper published today in the Proceedings of the National Academy of Sciences. “What I want to emphasize about this vaccine strategy is that it is broad,” said UCR virologist and paper author Rong Hai. “It is broadly applicable to any number of viruses, broadly effective against any variant of a virus, and safe for a broad spectrum of people. This could be the universal vaccine that we have been looking for.”
Continue Reading.
22K notes
·
View notes