#genetic sequencing
Text
5 notes
·
View notes
Text
First, let’s address the fact that hackers recently accessed the personal data of about 14,000 23andMe customers. Because of how 23andMe works—it has a “DNA Relatives” feature that lets users find people they are probably related to—this breach created 6.9 million “other users” who had data stolen in the breach, according to reporting by TechCrunch. This data included people’s names, birth year, relationships, percentage of DNA shared with other 23andMe users, and ancestry reports.
[...]
Getting your DNA or your loved ones’ DNA sequenced means you are potentially putting people who are related to those people at risk in ways that are easily predictable, but also in ways we cannot yet predict because these databases are still relatively new. I am writing this article right now because of the hack, but my stance on this issue has been the same for years, for reasons outside of the hack.
In 2016, I moderated a panel at SXSW called “Is Your Biological Data Safe?,” which was broadly about the privacy implications of companies and other entities creating gigantic databases of people’s genetic code. This panel’s experts included a 23andMe executive as well as an FBI field agent. Everyone on the panel and everyone in the industry agrees that genetic information is potentially very sensitive, and the use of DNA to solve crimes is obviously well established.
At the time, many of the possible dangers of providing your genome to a DNA sequencing company were hypothetical. Since then, many of the hypothetical issues we discussed have become a reality in one way or another. For example, on that panel, we discussed the work of an artist who was turning lost strands of hair, wads of chewing gum, and other found DNA into visual genetic “portraits” of people. Last year, the Edmonton Police Service, using a company called Parabon, used a similar process to create 3D images of crime suspects using DNA from the case. The police had no idea if the portrait they generated actually looked like the suspect they wanted, and the practice is incredibly concerning.
To its credit, 23andMe itself has steadfastly resisted law enforcement requests for information, but other large databases of genetic information have been used to solve crimes. Both 23andMe and Ancestry are regularly the recipients of law enforcement requests for data, meaning police do see these companies as potentially valuable data mines.
753 notes
·
View notes
Link
The sequencing and assembly of the human Y chromosome have been challenging due to its intricate repeat structure, which encompasses lengthy palindromes, tandem repeats, and segmental duplications. The existing GRCh38 reference sequence lacks over half of the Y chromosome’s content, leaving it as the final unfinished human chromosome. Addressing these limitations, the Telomere-to-Telomere (T2T) consortium introduces the full 62,460,029-base-pair sequence of the human Y chromosome from the HG002 genome (T2T-Y). This new sequence rectifies multiple errors in GRCh38-Y, supplementing the reference with over 30 million base pairs. It reveals complete ampliconic patterns of gene families like TSPY, DAZ, and RBMY, adds 41 protein-coding genes (primarily from TSPY), and unveils a distinctive alternating arrangement of human satellite 1 and 3 blocks within the heterochromatic Yq12 region. Through integration with the CHM13 genome assembly and the inclusion of population variations, clinical variants, and functional genomics data, a comprehensive reference spanning all 24 human chromosomes has been established.
The complex architecture of the human Y chromosome, with its large repeats and palindromes, plays a pivotal role in fertility. This includes hosting genes that are crucial for spermatogenesis and sex determination. Although it remains notably incomplete due to over half of its makeup being riddled with gaps in the GRCh38 human reference genome, this condition impedes comprehensive analysis of regions such as Azoospermia factors associated with infertility.
Despite these challenges, breakthroughs from the Telomere-to-Telomere (T2T) consortium have allowed researcher Adam M. Phillippy and his team to assemble the entire CHM13 cell line genome; however, they could not fully put together the Y chromosome because of its unique characteristics. The Human Pangenome Reference Consortium (HPRC) concurrently launched a project to embody a broader genomic range via the use of the HG002 genome. This endeavor successfully resulted in the reconstruction of the full sequence for the Y chromosome, known as T2T-Y.
Continue Reading
63 notes
·
View notes
Text
HOW DARE YOU FUK MY BGI™ Revolocity™ DNA sequencer© HE'S A VERY GOOD BOY and provides 50x faster whole genome sequences at 50X mean coverage, high quality SNP, CNV, Indel and SV data sequencing in standard file formats, comprehensive bioinformatics included!!! HE IS NOT SEXY


ok maybe a little
#eroticism of the machine#BGI™ Revolocity™ System DNA sequencer#science#genetics#biology#mad scientist
33 notes
·
View notes
Text

Exploring RNA Interference
Imagine a molecular switch within your cells, one that can selectively turn off the production of specific proteins. This isn't science fiction; it's the power of RNA interference (RNAi), a groundbreaking biological process that has revolutionized our understanding of gene expression and holds immense potential for medicine and beyond.
The discovery of RNAi, like many scientific breakthroughs, was serendipitous. In the 1990s, Andrew Fire and Craig Mello were studying gene expression in the humble roundworm, Caenorhabditis elegans (a tiny worm). While injecting worms with DNA to study a specific gene, they observed an unexpected silencing effect - not just in the injected cells, but throughout the organism. This puzzling phenomenon, initially named "co-suppression," was later recognized as a universal mechanism: RNAi.
Their groundbreaking work, awarded the Nobel Prize in 2006, sparked a scientific revolution. Researchers delved deeper, unveiling the intricate choreography of RNAi. Double-stranded RNA molecules, the key players, bind to a protein complex called RISC (RNA-induced silencing complex). RISC, equipped with an "Argonaut" enzyme, acts as a molecular matchmaker, pairing the incoming RNA with its target messenger RNA (mRNA) - the blueprint for protein production. This recognition triggers the cleavage of the target mRNA, effectively silencing the corresponding gene.
So, how exactly does RNAi silence genes? Imagine a bustling factory where DNA blueprints are used to build protein machines. RNAi acts like a tiny conductor, wielding double-stranded RNA molecules as batons. These batons bind to specific messenger RNA (mRNA) molecules, the blueprints for proteins. Now comes the clever part: with the mRNA "marked," special molecular machines chop it up, effectively preventing protein production. This targeted silencing allows scientists to turn down the volume of specific genes, observing the resulting effects and understanding their roles in health and disease.
The intricate dance of RNAi involves several key players:dsRNA: The conductor, a long molecule with two complementary strands. Dicer: The technician, an enzyme that chops dsRNA into small interfering RNAs (siRNAs), about 20-25 nucleotides long. RNA-induced silencing complex (RISC): The ensemble, containing Argonaute proteins and the siRNA. Target mRNA: The specific "instrument" to be silenced, carrying the genetic instructions for protein synthesis.
The siRNA within RISC identifies and binds to the complementary sequence on the target mRNA. This binding triggers either:Direct cleavage: Argonaute acts like a molecular scissors, severing the mRNA, preventing protein production. Translation inhibition: RISC recruits other proteins that block ribosomes from translating the mRNA into a protein.
From Labs to Life: The Diverse Applications of RNAi
The ability to silence genes with high specificity unlocks various applications across different fields:
Unlocking Gene Function: Researchers use RNAi to study gene function in various organisms, from model systems like fruit flies to complex human cells. Silencing specific genes reveals their roles in development, disease, and other biological processes.
Therapeutic Potential: RNAi holds immense promise for treating various diseases. siRNA-based drugs are being developed to target genes involved in cancer, viral infections, neurodegenerative diseases, and more. Several clinical trials are underway, showcasing the potential for personalized medicine.
Crop Improvement: In agriculture, RNAi offers sustainable solutions for pest control and crop development. Silencing genes in insects can create pest-resistant crops, while altering plant genes can improve yield, nutritional value, and stress tolerance.
Beyond the Obvious: RNAi applications extend beyond these core areas. It's being explored for gene therapy, stem cell research, and functional genomics, pushing the boundaries of scientific exploration.
Despite its exciting potential, RNAi raises ethical concerns. Off-target effects, unintended silencing of non-target genes, and potential environmental risks need careful consideration. Open and responsible research, coupled with public discourse, is crucial to ensure we harness this powerful tool for good.
RNAi, a testament to biological elegance, has revolutionized our understanding of gene regulation and holds immense potential for transforming various fields. As advancements continue, the future of RNAi seems bright, promising to silence not just genes, but also diseases, food insecurity, and limitations in scientific exploration. The symphony of life, once thought unchangeable, now echoes with the possibility of fine-tuning its notes, thanks to the power of RNA interference.
#science sculpt#life science#science#molecular biology#biology#biotechnology#dna#double helix#genetics#artists on tumblr#rna#rna sequencing#RNA interference#cell biology#cells#biomolecules#illustrates#scientific illustration#illustration#illustrative art#scientific research
12 notes
·
View notes
Text

i am literally always saying this
9 notes
·
View notes
Note
Hi! You mentioned in your pinned post your studying biochemical engineering and computer science, and I'd like to ask:
Why did you choose those? (if it's not too personal)
and
What's your favorite little quirk/big quirk/whatever about biochemistry and/or computer science?
hi! sorry this took me like a century to answer :p
so i chose those two through a process of elimination. before i even got to college i knew that i wanted to study STEM, but i had no idea which specific field i wanted to go into. chemistry always sounded the most fun to me, and i like working with computers, so i started with those. i took a few classes in various STEM fields and talked to some people at my university which helped me narrow down my interests. i took chemistry for a year and half and i liked it a lot, but i wanted to try biology, so i changed my major to evolutionary genetics. after studying primarily biology for a semester, i decided it wasn't really my thing, but i still enjoyed certain aspects of it: mainly protein function within an organism and the chemical interactions of our cells. so i switched to biochemistry. my computer science interest is a bit more straightforward: computers are cool, and knowledge of them is useful in a wide variety of fields. i figure it's a good skill to have and this way, it's not necessarily an either-or thing (which universities are very big on).
as for "quirks," i'm not entirely sure how you mean, but i really like studying enantiomeric resolution and substitution/elimination reactions. it's been a little while since i've done it but i think it's cool how you can use molecular and electron geometry to change the solubility of otherwise identical compounds and retrieve one from a racemic mixture, and i'm excited to learn about the potential applications in a biological system.
for comp sci, i haven't been studying it as much as i have natural sciences, but one thing i thought was helpful conceptually was this:
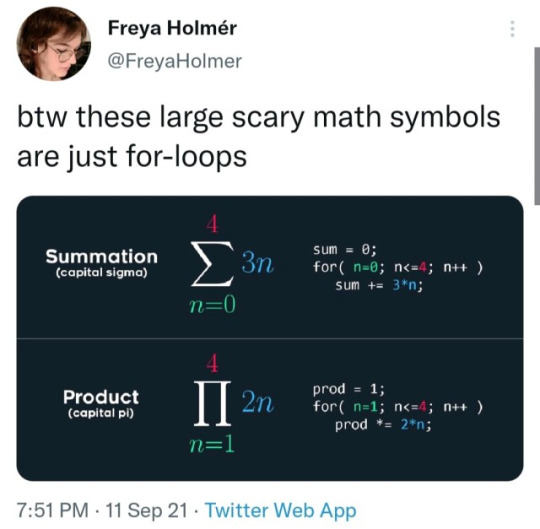
#but yeah i might go into vaccine development after getting my degree#ive also heard from some academic advisors that computer science is useful if i decide to go into genetics after all#because they can more easily program gene sequences#inbox
22 notes
·
View notes
Text
When a computer slows down bc its processing a shit ton of sequencing data i say "gotta be patient, the computer is doing a big thinkie"
#genetics#sequencing#RNA sequencing#scRNA#single cell sequencing#genomics#this computer has 4 cores and idk how much fuckin ram#but damn if it still wont take 20 min to do the alignments#of just ONE sample#also that's just one step >.<#counts take a while too
14 notes
·
View notes
Text
absolutely hilarious that I instantly went to the tag “largo siblings” (shocking to no one who has ever met me or had a conversation with me) & of course I came to some boring predictable long ass post about how it’s sick & twisted & misinterprets the movie to think the largos are sympathetic.
& of course I was like “pshhhh lmao I highly doubt anyone is actually saying the largos are sympathetic that’s so stupid this person definitely misinterpreted people just having fun & liking villains on purpose”
…. & like a few posts down someone was talking about how they sympathize with the largos & want them to be good people. this website is so predictable & yet I’m somehow baffled every time.
anyway the largo siblings are hot & id let them fuck up my life
(& I shouldn’t have to clarify this but yes obviously i understood the very obvious & not subtle commentary on wealth healthcare & exploitation of the movie)
#I just think that sequence of events was really really comedic & predictable#OBVIOUSLY I went to the tag for the evil fucked up siblings if u didn’t call that one I don’t think you’ve ever interacted with me once#& obviously I rolled my eyes at the person explaining obvious stuff & just assumed they were making up a guy to be mad at#& then obviously they really were not people are actually that stupid#I don’t know why I think that whole sequence of events was so hilarious but it was. to me#largo siblings#repo! the genetic opera#movie tag
4 notes
·
View notes
Text
tbh i think that. yeah the most challenging pokemon side games that still are hard af as an adult are conquest and ranger. like those games pull no punches.
#THE WAY RANGER CAN FUCK YOU OVER#ESPECIALLY OG RANGER#WHERE YOU HAD TO LITERALLY DO EVERY LOOP IN A ROW OR IT WOULD RESET YOUR COUNTER#BEATING THE FINAL BOSS ENTEI I WAS CLENCHING#iirc the final boss entei you literally had to loop that bitch uninterrupted like 40 times or something like that to capture it#test of fortitude#at least all the partner pokemon in the series are extremely helpful#i loved the multiple partners choices of soa#i still am so chuffed when i befriend an adorable pachirisu or kricketot and they actually really pull their weight in a game for once#ukulele pichu however will always remain the most broken partner#the fucking ukelele attack getting even stronger as you progress to the point you just paralyze everything is so fun#and tbh the moment i figured out that you could use the fucking symbol in the INTRO SEQUENCE to summon a FREE MEWTWO#that was SO COOL#i would just summon him and be buddies with him i rarely ever used him#just my buddy Genetic Freak#hades.txt
4 notes
·
View notes
Text
The White House has issued new rules aimed at companies that manufacture synthetic DNA after years of warnings that a pathogen made with mail-order genetic material could accidentally or intentionally spark the next pandemic.
The rules, released on April 29, are the result of an executive order signed by President Joe Biden last fall to establish new standards for AI safety and security, including AI applied to biotechnology.
Artificially generated DNA allows researchers to do all sorts of things—develop diagnostic tests, make beneficial enzymes to eat up plastic, or engineer potent antibodies to treat disease—without having to extract natural sequences from organisms. Need to study a rare type of bacteria? Instead of going out into the field to collect a sample, its genetic sequence can simply be ordered from a DNA synthesis company instead.
Synthesizing DNA has been possible for decades, but it’s become increasingly easier, cheaper, and faster to do so in recent years thanks to new technology that can “print” custom gene sequences. Now, dozens of companies around the world make and ship synthetic nucleic acids en masse. And with AI, it’s becoming possible to create entirely new sequences that don’t exist in nature—including those that could pose a threat to humans or other living things.
“The concern has been for some time that as gene synthesis has gotten better and cheaper, and as more companies appear and more technologies streamline the synthesis of nucleic acids, that it is possible to de novo create organisms, particularly viruses,” says Tom Inglesby, an epidemiologist and director of the Johns Hopkins Center for Health Security.
It’s conceivable that a bad actor could make a dangerous virus from scratch by ordering its genetic building blocks and assembling them into a whole pathogen. In 2017, Canadian researchers revealed they had reconstructed the extinct horsepox virus for $100,000 using mail-order DNA, raising the possibility that the same could be done for smallpox, a deadly disease that was eradicated in 1980.
The new rules aim to prevent a similar scenario. It asks DNA manufacturers to screen purchase orders to flag so-called sequences of concern and assess customer legitimacy. Sequences of concern are those that contribute to an organism’s toxicity or ability to cause disease. For now, the rules only apply to scientists or companies that receive federal funding: They must order synthetic nucleic acids from providers that implement these practices.
Inglesby says it’s still a “big step forward” since about three-quarters of the US customer base for synthetic DNA are federally funded entities. But it means that scientists or organizations with private sources of funding aren’t beholden to using companies with these screening procedures.
Many DNA providers already follow screening guidelines issued by the Department of Health and Human Services in 2010. About 80 percent of the industry has joined the International Gene Synthesis Consortium, which pledges to vet orders. But these measures are both voluntary, and not all companies comply.
Kevin Flyangolts, founder and CEO of New York–based Aclid, a company that offers screening software to DNA providers, says he’s glad to see the White House taking action. “While the industry has done a pretty good job of putting some protocols in place, it’s by and large not consistent,” he says. Still, he hopes Congress will adopt formal legislation by requiring all DNA providers to screen orders.
Last year, a bipartisan group of legislators introduced the Securing Gene Synthesis Act to mandate screening more broadly, but the bill has yet to advance.
Emily Leproust, CEO of Twist Bioscience, a San Francisco DNA-synthesis company, welcomes regulation. “We recognize that DNA is dual-use technology. It’s like dynamite, you can build tunnels, but you can also kill people,” she says. “Collectively, we have a responsibility to promote the ethical use of DNA.”
Twist has been screening sequences and customers since 2016, when it first started selling nucleic acids to customers. A few years ago, the company hired outside consultants to test its screening processes. The consultants set up fake customer names and surreptitiously ordered sequences of concern.
Leproust says the company successfully flagged many of those orders, but in some cases, there was internal disagreement on whether the sequences requested were worrisome or not. The exercise helped Twist adopt new protocols. For instance, it used to only screen DNA sequences 200 base pairs or longer. (A base pair is a unit of two DNA letters that pair together.) Now, it screens ones that are at least 50 base pairs to prevent customers from shopping around for smaller sequences to assemble together.
While Twist has tightened its own screening measures, Leproust still worries about some hypothetical scenarios that are beyond her control. For instance, a state actor with bad intentions could start making its own gene sequences. “Probably the biggest risk is if a state wants to build their own DNA synthesis capabilities,” she says. “They may be able to do it, because states have vast resources.”
#us politics#news#wired#biden administration#president joe biden#2024#synthetic DNA#regulations#International Gene Synthesis Consortium#Department of Health and Human Services#Johns Hopkins Center for Health Security#Securing Gene Synthesis Act#dna#science#Twist Bioscience#Aclid#bioscience#genetics#diseases#pathogens#bacteria#custom gene sequences#gene synthesis
2 notes
·
View notes
Text
Maximizing Efficiency: Best Practices for Using Sequencing Consumables

By implementing these best practices, researchers can streamline sequencing workflows, increase throughput, and achieve more consistent and reproducible results in genetic research. Sequencing Consumables play a crucial role in genetic research, facilitating the preparation, sequencing, and analysis of DNA samples. To achieve optimal results and maximize efficiency in sequencing workflows, it's essential to implement best practices for using these consumables effectively.
Proper planning and organization are essential for maximizing efficiency when using Sequencing Consumables. Before starting a sequencing experiment, take the time to carefully plan out the workflow, including sample preparation, library construction, sequencing runs, and data analysis. Ensure that all necessary consumables, reagents, and equipment are readily available and properly labeled to minimize disruptions and delays during the experiment.
Optimizing sample preparation workflows is critical for maximizing efficiency in sequencing experiments. When working with Sequencing Consumables for sample preparation, follow manufacturer protocols and recommendations closely to ensure consistent and reproducible results. Use high-quality consumables and reagents, and perform regular quality control checks to monitor the performance of the workflow and identify any potential issues early on.
Utilizing automation technologies can significantly increase efficiency when working with Sequencing Consumables. Automated sample preparation systems and liquid handling robots can streamline repetitive tasks, reduce human error, and increase throughput. By automating sample processing and library construction workflows, researchers can save time and resources while improving consistency and reproducibility in sequencing experiments.
Get More Insights On This Topic: Sequencing Consumables
#Sequencing Consumables#DNA Sequencing#Laboratory Supplies#Genetic Analysis#Next-Generation Sequencing#Molecular Biology#Research Tools#Bioinformatics
2 notes
·
View notes
Text

Paper tissues white as snow come away bloody, the consistency of what she dabs away being so dark and thick it's more like tar than blood. Gradually her coughing fit recedes, lungs burning like fire as the flow of oxygen resumes though it's obvious from the way she looks that Khare's not feeling any better. She's not well, eyes dull and glassy as she tiredly cleans herself up. "M'sorry about that. It's... not contagious, promise."
#🌈 || musings#;; IC Status#illness tw#illness cw#Just a smol to get myself in the writing mood again this weekend!#Khare's not well but I'm feeling great lol#She gets coughing fits like this from time to time#Happens more frequently the more her mutation advances#Your whole genetic sequence just falling to bits kinda type deal situation you know :')#Just take a night off work my girl#If you don't like this you're gonna hate what's coming asjsjdg#Hope you're all having a great weekend lovelies!
10 notes
·
View notes
Link
In a monumental milestone, UK Biobank has unveiled open access to incredible whole genome sequencing data for its over half a million participants. This vast genomic resource promises to massively accelerate the diagnosis, treatment, and prevention of diseases worldwide when combined with the study’s expansive health data. Dubbed a “veritable treasure trove” by Professor Sir Rory Collins, Principal Investigator, UK Biobank, the release cements the nonprofit UK Biobank as the most ambitious health research undertaking ever.
Genomic sequencing has revolutionized biomedicine by enabling the reading of the precise DNA code underlying human life. Technologies like PCR unlocked targeted gene testing decades ago. However, the affordable sequencing of entire genomes on a mass scale has only recently become feasible.
While costs have plunged, interpreting genomic data relies heavily on computational analytics. UK Biobank’s dataset amalgamates sequenced genomes from over 350,000 British volunteers with intricate health details like brain imaging, blood assays, lifestyle surveys, and more collected meticulously for 15 years. This unrivaled fusion promises tremendous leaps in illuminating genetics‘ role in disease.
Continue Reading
50 notes
·
View notes
Text
I've been having some pretty strong disability feelings lately.
I get why the push to discretize EDS was made and I honestly agree with it, especially in outlining the different genetic links as a part of the diagnostic criteria. It really had/has become a "problems disorder to stick people in when they have these general problems I guess", which medically makes it more difficult to treat than necessary. By laying out the boundaries for the thirteen types in 2017 each one becomes easier to treat (not to be confused with easy to treat!), and more consistent to describe. I agree with those choices.
I, personally, just wish I could've gone through the process just a little bit earlier. My diagnostic ~journey~ started in like 2012 when I couldn't push past my knees being stupid any longer, but I didn't get properly diagnosed and treated until I turned 18 and spent the next three years aggressively chasing specialists and advocating for myself. I got genetic testing in 2019, and don't have any of the known genetic markers. Everyone I've spoken to who knows about these things has done a double take at that, and asked if I'm *sure* I don't have classical or vascular. The geneticist herself looked at me and said if it were a couple of years earlier she'd just diagnose me with classical on the spot and send me back to cardio to test for vascular.
So whatever, I'm not in the genetic club that's being tracked at the moment, boo hoo just cope with everyone else in the HSD/hEDS bucket.
the reason I'm so frustrated is because this clearly runs in my family, my specific symptomology and presentation. aortic dissection has killed two of the last seven people to die in my blood family! that is ten times the average incidence! we don't age/have that eds baby face forever! the only reason I haven't had any of my freaky under-skin lumps analyzed is because both my mother and my grandmother have had theirs biopsied to hell and back and just get a shrug emoji! my little brother has the same joint problems I started presenting with and I can't help but hope both that his don't progress and that they do so I won't be the only moderate case we know of antemortem! I may be the only one of us living who has presented this severely but I've also been under enough stress to activate every epigenetic problem I've got! fuck!
this post brought to you by my family's selective breeding program
#heds#long post#disability rant#i am tired and i am tired of ehlers danlos syndrome specifically#and I wish I had the money and the connections to bring a vial of blood from all my living relatives and demand it sequenced and analyzed#for some validation#so I dont feel so fucking crazy for being so functional and disfunctional at once#aiyi. sometimes diversifying the gene pool for your progeny is a great plan and sometimes you invent cool new genetic disorders ig
3 notes
·
View notes
Text
"With the announcement that the Human Genome Project had mapped all of the genetic material in the human chromosomes, a new era in the understanding of genetics began. The discovery of new genes is announced every day, and it is only a matter of time before the genetic mechanisms of mood disorders is only one of the goals of work in this field. Just as important will be understanding the epigenetic mechanisms by which genes turn on and off and other mechanisms that regulate the expression and work of the instructions encoded in the DNA molecule.
The first genetic approach to pay off in changing treatment is likely to be pharmacogenomics, the field within genetics that investigates genetic factors associated with responses to particular pharmaceuticals rather than with risk of disease. The promise of pharmacogenomics is that therapeutic agents can be rationally selected, based on a person's genetic profile rather than the trial-and-error process patients must now endure. In the not-too-distant future, a blood test will show whether lithium or valproate or lamotrigine or some as yet undiscovered drug will be the best treatment for a particular individual with bipolar disorder. A blood test may be able to identify the bipolar patients who can safely take an antidepressant."
-Francis Mark Mondimore, Bipolar Disorder (2014)
#books#quote#nonfiction#bipolar disorder#mental health#psychology#genomics#dna sequencing#dna#genetics#science#biology
3 notes
·
View notes