#cell and molecular biology
Text
5 notes
·
View notes
Text
"A team at Northwestern University has come up with the term “dancing molecules” to describe an invention of synthetic nanofibers which they say have the potential to quicken the regeneration of cartilage damage beyond what our body is capable of.
The moniker was coined back in November 2021, when the same team introduced an injection of these molecules to repair tissues and reverse paralysis after severe spinal cord injuries in mice.
Now they’ve applied the same therapeutic strategy to damaged human cartilage cells. In a new study, published in the Journal of the American Chemical Society, the treatment activated the gene expression necessary to regenerate cartilage within just four hours.
And, after only three days, the human cells produced protein components needed for cartilage regeneration, something humans can’t do in adulthood.
The conceptual mechanisms of the dancing molecules work through cellular receptors located on the exterior of the cell membrane. These receptors are the gateways for thousands of compounds that run a myriad of processes in biology, but they exist in dense crowds constantly moving about on the cell membrane.
The dancing molecules quickly form synthetic nanofibers that move according to their chemical structure. They mimic the extracellular matrix of the surrounding tissue, and by ‘dancing’ these fibers can keep up with the movement of the cell receptors. By adding biological signaling receptors, the whole assemblage can functionally move and communicate with cells like natural biology.
“Cellular receptors constantly move around,” said Northwestern Professor of Materials Sciences Samuel Stupp, who led the study. “By making our molecules move, ‘dance’ or even leap temporarily out of these structures, known as supramolecular polymers, they are able to connect more effectively with receptors.”
The target of their work is the nearly 530 million people around the globe living with osteoarthritis, a degenerative disease in which tissues in joints break down over time, resulting in one of the most common forms of morbidity and disability.
“Current treatments aim to slow disease progression or postpone inevitable joint replacement,” Stupp said. “There are no regenerative options because humans do not have an inherent capacity to regenerate cartilage in adulthood.”
In the new study, Stupp and his team looked to the receptors for a specific protein critical for cartilage formation and maintenance. To target this receptor, the team developed a new circular peptide that mimics the bioactive signal of the protein, which is called transforming growth factor beta-1 (TGFb-1).
Northwestern U. Press then reported that the researchers incorporated this peptide into two different molecules that interact to form supramolecular polymers in water, each with the same ability to mimic TGFb-1...
“With the success of the study in human cartilage cells, we predict that cartilage regeneration will be greatly enhanced when used in highly translational pre-clinical models,” Stupp said. “It should develop into a novel bioactive material for regeneration of cartilage tissue in joints.”
“We are beginning to see the tremendous breadth of conditions that this fundamental discovery on ‘dancing molecules’ could apply to,” Stupp said. “Controlling supramolecular motion through chemical design appears to be a powerful tool to increase efficacy for a range of regenerative therapies.”"
-via Good News Network, August 5, 2024
#nanotechnology#osteoarthritis#arthritis#medical news#science news#cell biology#molecular biology#cartilage#good news#hope
704 notes
·
View notes
Text
As direct descendants of ancient bacteria, mitochondria have always been a little alien. Now a study shows that mitochondria are possibly even stranger than we thought.
The study, titled "Somatic nuclear mitochondrial DNA insertions are prevalent in the human brain and accumulate over time in fibroblasts," appears in PLOS Biology.
Mitochondria in our brain cells frequently fling their DNA into the nucleus, the study found, where the DNA becomes integrated into the cells' chromosomes. And these insertions may be causing harm: Among the study's nearly 1,200 participants, those with more mitochondrial DNA insertions in their brain cells were more likely to die earlier than those with fewer insertions.
Continue Reading.
149 notes
·
View notes
Text
I was in bio the other week and thinking about mitochondria (as you do) and I kept thinking about a heavy metal poster that was mitochondria and like

I did it lol
#u can get this on my Redbubble btw!!#mitochondria#mitochondria is the powerhouse of the cell#biology#bio#microbiology#cells#molecular biology#cell biology
321 notes
·
View notes
Text


I am still stuck on the same data analysis (but finally towards the end !). To avoid posting the same desk pic for days in a row, I'll have you meet my lil study buddy 🥰
#studyblr#studyspo#my post#original post#study motivation#biology major#molecular biology#chaotic academia#academics#study desk#study buddy#biology student#cell biology#biology#study blog#desk aesthetic#bacteria#bacteriology#biomedical science#sylvanian families#stem studyblr#stem academia#stemblr#women in stem#stem student
26 notes
·
View notes
Text

Plant Immune System Part 3
The plant immune system is the topic of my PhD thesis, which I'm currently writing following several years of lab-based research as a PhD student at Imperial College London under the supervision of Professor Colin Turnbull.
Here's an introduction to my research, which focused on how certain plants defend themselves against aphids.
Aphids are an important insect pest that threaten agriculture worldwide. As we learned in the previous post, plant resistance (R) genes control resistance to specific pests and pathogens through interaction with effectors from the invaders. Since examples of R gene-dependent aphid resistance have been documented in different plant species, aphid-specific R genes may enable the development of resistant crops.
In the model plant Medicago truncatula, there are some varieties that are resistant to aphids and other varieties that are susceptible to Pea Aphids (Acyrthosiphon pisum). Whether the plant is resistant also depends on the variety of aphid. In my project, the A17 plant is resistant to PS01 aphids but not to N116 aphids, while the DZA plant is susceptible to both aphid varieties.
What is the key difference in the resistant versus susceptible plants? Resistant A17 plants have a portion of their genome “Resistance to Acyrthosiphon pisum 1” (RAP1) which determines resistance to PS01 aphids, but the genes controlling the defence response and physiological defence mechanisms remain unknown. Two candidate R genes located in RAP1, designated “RAP1A” and “RAP1B”, may control resistance.
My main objective in my PhD project has been to determine whether RAP1A and RAP1B control aphid resistance, and to investigate the RAP1-mediated defence response. I look forward to sharing the findings in publications and in talks next year!
Image credit: Original diagram by Katia Hougaard with images from the Turnbull Lab.
#katia_plantscientist#science#biology#research#plants#botany#plantbiology#phdproject#plantbiology#plantscience#sciencecommunication#diagrams#phd#imperialcollegelondon#phdthesis#medicago#aphid#plantimmunesystem#pestsandpathogens#plantpathology#womeninscience#plantbiologist
#katia plant scientist#botany#plant biology#plants#plant science#diagram#science communication#science#sciencecore#plant scientist#biology#biologist#phdblr#phd life#phdjourney#phd student#phd research#gradblr#stem#immune system#cell biology#molecular biology#genetics#aphid#medicago#clover#plant pathology#my work
12 notes
·
View notes
Text
Finals Week is here, and ya know what that means? Ikemen Vampire has found its way into my studying once again.





This really does help me remember stuff better. Highly recommend it if you find it works for you. It also just makes studying more fun.
#ikemen vampire#cybird ikemen#ikemenvamp#ikevamp#studying#studying but with anime#rice rambles#college life#study tips#molecular and cell biology
29 notes
·
View notes
Text
Cat Color Basics: Eumelanin and Phaeomelanin
In mammals, there are a type of cells located in the skin called melanocytes. These cells produce a pigment called melanin which is responsible for the color of their skin and fur. There are two types of melanin; eumelanin and phaeomelanin. Eumelanin is black and brown pigment, while phaeomelenin is red pigment. The wide variety in the color of cats that we see are an outcome from mutations in the genes that result in modifications of pigment production, granule placement, and more.
Eumelanin-based colors

Phaeomelanin-based colors

Note: Breeders may call these colors by other names, such as black being labeled ebony
#genetics#cat genetics#cat colors#biology#molecular biology#cell biology#science#cats#cat art#cat drawing
11 notes
·
View notes
Text
i love lecture recordings, i genuinely love listening to these like podcasts. this is entertainment to me, fun, learning in a way that does not bore me!
the moment they give me a single 10 page article though it takes me 5 business weeks to read it
#why are all audiobooks fiction or pop science at best. i want to listen to my textbooks#molecular biology of the cell 4th edition audiobook when. fundamentals of pharmaceutical chemistry audiobook WHEN
52 notes
·
View notes
Text
”You matter” no please, I find my insignificance quite comforting, actually.
#oh to just be a tiny pebble in a stream#free of problems#yes this is about my chemistry group assignment#i have no idea what i’m doing here#I’ve been studying chemistry for 8 semesters why do they want us to do molecular biology again#i am developing a passionate hate for cell assays#stem student#university life#i guess
7 notes
·
View notes
Text

Exploring RNA Interference
Imagine a molecular switch within your cells, one that can selectively turn off the production of specific proteins. This isn't science fiction; it's the power of RNA interference (RNAi), a groundbreaking biological process that has revolutionized our understanding of gene expression and holds immense potential for medicine and beyond.
The discovery of RNAi, like many scientific breakthroughs, was serendipitous. In the 1990s, Andrew Fire and Craig Mello were studying gene expression in the humble roundworm, Caenorhabditis elegans (a tiny worm). While injecting worms with DNA to study a specific gene, they observed an unexpected silencing effect - not just in the injected cells, but throughout the organism. This puzzling phenomenon, initially named "co-suppression," was later recognized as a universal mechanism: RNAi.
Their groundbreaking work, awarded the Nobel Prize in 2006, sparked a scientific revolution. Researchers delved deeper, unveiling the intricate choreography of RNAi. Double-stranded RNA molecules, the key players, bind to a protein complex called RISC (RNA-induced silencing complex). RISC, equipped with an "Argonaut" enzyme, acts as a molecular matchmaker, pairing the incoming RNA with its target messenger RNA (mRNA) - the blueprint for protein production. This recognition triggers the cleavage of the target mRNA, effectively silencing the corresponding gene.
So, how exactly does RNAi silence genes? Imagine a bustling factory where DNA blueprints are used to build protein machines. RNAi acts like a tiny conductor, wielding double-stranded RNA molecules as batons. These batons bind to specific messenger RNA (mRNA) molecules, the blueprints for proteins. Now comes the clever part: with the mRNA "marked," special molecular machines chop it up, effectively preventing protein production. This targeted silencing allows scientists to turn down the volume of specific genes, observing the resulting effects and understanding their roles in health and disease.
The intricate dance of RNAi involves several key players:dsRNA: The conductor, a long molecule with two complementary strands. Dicer: The technician, an enzyme that chops dsRNA into small interfering RNAs (siRNAs), about 20-25 nucleotides long. RNA-induced silencing complex (RISC): The ensemble, containing Argonaute proteins and the siRNA. Target mRNA: The specific "instrument" to be silenced, carrying the genetic instructions for protein synthesis.
The siRNA within RISC identifies and binds to the complementary sequence on the target mRNA. This binding triggers either:Direct cleavage: Argonaute acts like a molecular scissors, severing the mRNA, preventing protein production. Translation inhibition: RISC recruits other proteins that block ribosomes from translating the mRNA into a protein.
From Labs to Life: The Diverse Applications of RNAi
The ability to silence genes with high specificity unlocks various applications across different fields:
Unlocking Gene Function: Researchers use RNAi to study gene function in various organisms, from model systems like fruit flies to complex human cells. Silencing specific genes reveals their roles in development, disease, and other biological processes.
Therapeutic Potential: RNAi holds immense promise for treating various diseases. siRNA-based drugs are being developed to target genes involved in cancer, viral infections, neurodegenerative diseases, and more. Several clinical trials are underway, showcasing the potential for personalized medicine.
Crop Improvement: In agriculture, RNAi offers sustainable solutions for pest control and crop development. Silencing genes in insects can create pest-resistant crops, while altering plant genes can improve yield, nutritional value, and stress tolerance.
Beyond the Obvious: RNAi applications extend beyond these core areas. It's being explored for gene therapy, stem cell research, and functional genomics, pushing the boundaries of scientific exploration.
Despite its exciting potential, RNAi raises ethical concerns. Off-target effects, unintended silencing of non-target genes, and potential environmental risks need careful consideration. Open and responsible research, coupled with public discourse, is crucial to ensure we harness this powerful tool for good.
RNAi, a testament to biological elegance, has revolutionized our understanding of gene regulation and holds immense potential for transforming various fields. As advancements continue, the future of RNAi seems bright, promising to silence not just genes, but also diseases, food insecurity, and limitations in scientific exploration. The symphony of life, once thought unchangeable, now echoes with the possibility of fine-tuning its notes, thanks to the power of RNA interference.
#science sculpt#life science#science#molecular biology#biology#biotechnology#dna#double helix#genetics#artists on tumblr#rna#rna sequencing#RNA interference#cell biology#cells#biomolecules#illustrates#scientific illustration#illustration#illustrative art#scientific research
12 notes
·
View notes
Text


brain organoid culture
3 notes
·
View notes
Text

10 notes
·
View notes
Text
UCLA scientists have identified a protein that plays a critical role in regulating human blood stem cell self-renewal by helping them sense and interpret signals from their environment.
The study, published in Nature, brings researchers one step closer to developing methods to expand blood stem cells in a lab dish, which could make life-saving transplants of these cells more available and increase the safety of blood stem cell-based treatments, such as gene therapies.
Blood stem cells, also known as hematopoietic stem cells, have the ability to make copies of themselves via a process called self-renewal, and can differentiate to produce all the blood and immune cells found in the body. For decades, transplants of these cells have been used as life-saving treatments for blood cancers such as leukemia and various other blood and immune disorders.
Continue Reading.
197 notes
·
View notes
Text
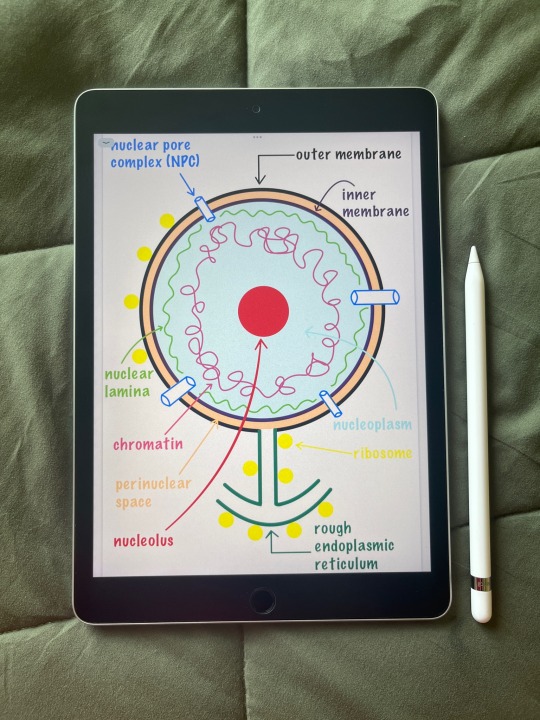
studying cell biology brings me so much joy fr 🦠
25 notes
·
View notes
Text



snapshots of last week ! send strength for the experiments of the upcoming days 😪
#studyblr#studyspo#my post#original post#study motivation#chaotic academia#academics#biology major#molecular biology#cell biology#bacteria#dark academia#stem studyblr#women in stem#stemblr#currently reading#books
13 notes
·
View notes