#RNA modification
Explore tagged Tumblr posts
Text
youtube
#m6A modification#28S rRNA#oncogenic mRNA translation#tyrosine catabolism#RNA epigenetics#ribosomal biology#cancer metabolism#tumor progression#mRNA regulation#epitranscriptomics#oncogene expression#translational control#metabolic pathways#RNA modification#ribosome recruitment#cancer research#mRNA stability#oncogenic signaling#molecular oncology#targeted cancer therapy.#Youtube
0 notes
Text
Can you imagine the laundry list of vaccines you'd need in a intergalactic society. They'd probably dole it out a little for most people but also have some kind of 100 ml Supreme Vaccine that gives you The Plague feat Extreme Arm Pain for a week straight and then inoculates you against absolutely everything.
#that or gene modification but that's not as fun#or maybe some RNA stuff#probably actually#this is mostly about losh but still#scifi thoughts
4 notes
·
View notes
Text
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra
Unlocking the Mysteries of Gene Expression: From Genomic Imprinting to Non-Coding RNAs in Biology Class with Dr. Mishra #GeneExpression #BiologyClass #GenomicImprinting #NonCodingRNAs #CollaborativeLearning
Dr. Mishra: “Hello there! I see we have a new face in our biology class today. Welcome! We’re so glad you’re here, and I want you to know that this classroom is a friendly and supportive place. If you ever have questions or need assistance, don’t hesitate to ask me or your classmates. We’re all here to learn together and make this an enjoyable experience for you.” Abby: “Hello. My name is Abha…

View On WordPress
#biology#Chromatin Structure#Collaborative Learning#Epigenetic Modifications#Gene expression#Genomic Imprinting#MicroRNAs#Non-Coding RNAs#Transcription Factors
0 notes
Text
All cells with mitochondria have a nucleus, all cells with a nucleus have mitochondria (or denegerated former mitochondria). It's not obvious that this should be so. In general, you should get branching after every trait (ofc this isn't always the case). If organisms with trait A are successful then there should be enough of them to branch. If they require trait B to be successful, trait A shouldn't have reached fixation in the first place, unless they were both caused by the same genetic modification.
Nick lanes theory, in the vital question, is that the mitochondria *directly* caused the development of the nucleus. If you're an archeon with bacteria living inside you, and one of them dies, it's membrane will dissolve and release its genetic material. You're an archeon, so you're used to doing lateral gene transfer, and will copy it's code into yours. This code has a poison and it's antidote
Bacterial genetic code has self-replicating parasitic genes. these genes are adapted to their bacterial host and splice themselves out before transcription. bacteria face strong selection to pare down their genome, so they dont have very many of these. but if you suddenly acquire a huge amount of bacterial genetic code, the parasites therein, not adapted to you, will put themselves in all sorts of bad places. then, because you dont face very strong selection, if these codes mutate in a way that breaks their ability to copy themselves, and splice themselves out before transcription, youll have a bunch of faulty genes. these dead regions are called introns. this is a huge problem! you can develop a protein to splice them out "manually", called the spliceosome, but it works slowly, too slowly to get them all fixed before they reach the ribosome to be made into proteins
HOWEVER, this bacterial code will also have a bunch of genetic code for bacterial membranes. the archeon will start producing a bunch of extra membrane enzymes, which will go around producing extra membranes. without adaptations to handle these, theyll just build up. around where theyre produced. lipids naturally form into closed surfaces in solution, so you'd end up with a bunch of lipid "bags" around your genome. but those bags are the solution to your intron problem! they impede the diffusion of the rna from the genome to the ribosome, giving the spliceosome time to work.
eventually (its theorized) these lipid bags evolved into an enclosed double membrane with pore membranes, but during mitosis they split into discrete lipid bags again!
95 notes
·
View notes
Text
Apparently McCoy speaks Vulcan and it can be learned there RNA modifications


93 notes
·
View notes
Text

Epigenetics: A Journey Through Inheritance Beyond Genes
For centuries, scientists have been fascinated by the mysteries of heredity and how traits are passed down from generation to generation. DNA, the molecule that stores our genetic code, was once thought to be the sole determinant of our characteristics. However, a new frontier in biology, revealing a captivating layer of complexity beyond the DNA sequence itself: Epigenetics.
What is Epigenetics?
The term "epigenetics" was first coined in the 1940s by British biologist Conrad Waddington, but it wasn't until the late 20th century that its significance truly blossomed. Epigenetics, literally meaning "above genetics," refers to the study of heritable changes in gene expression that occur without alterations to the DNA sequence itself. Imagine DNA as the musical score, but epigenetics are the conductor and musicians who determine how the music is played. Through chemical modifications and adjustments to the proteins around DNA, epigenetics dictates which genes are turned on or off, influencing how cells function and ultimately shaping our health, development, and even behavior. Think of your DNA as the hardware: it contains the basic instructions for building and running your body. But epigenetics acts like the software, fine-tuning those instructions and determining which genes get turned on or off at specific times and in specific cells. These modifications, like chemical tags or changes in the packaging of DNA, don't alter the underlying code itself, but they can have a profound impact on how it's read and interpreted.
The Key Players:
DNA methylation: This process involves adding a methyl group to DNA, essentially silencing the gene it's attached to. Imagine it like putting a dimmer switch on a light bulb.
Histone modifications: Histones are proteins that package DNA, and changes in their structure can make genes more or less accessible to the cellular machinery needed for expression. Think of it like adjusting the curtains around a window - open wide for full light, slightly closed for filtered light.
Non-coding RNAs: These are molecules that don't code for proteins but can regulate gene expression in various ways. They're like the backstage crew in a play, ensuring everything runs smoothly.
The Power of Epigenetic Regulation
Epigenetic regulation plays a crucial role in various biological processes, including:
Development: During embryonic development, different cell types emerge from the same DNA blueprint by activating or silencing specific gene sets through epigenetic modifications.
Cellular differentiation: Specialized cells like muscle or nerve cells have unique functions due to differences in their active genes, controlled by epigenetic mechanisms.
Learning and memory: Epigenetic changes in brain cells are thought to be essential for learning and forming memories.
Aging: As we age, our epigenome accumulates changes that can contribute to age-related decline and disease.
Environmental influences: Diet, exercise, stress, and exposure to toxins can leave epigenetic marks on our genes, potentially impacting our health and even the health of future generations.
Epigenetics reminds us that we are not simply products of our genes. Our environment, choices, and experiences leave their mark, shaping who we are and potentially influencing our children's health. This deeper understanding of ourselves opens doors for self-awareness, empowerment, and potentially reshaping our narratives – not just as individuals, but as a species with the potential to leave a healthier legacy for generations to come.
#life science#biology#science sculpt#molecular biology#biotechnology#epigenetics#daily dose of science#dna#genetic inheritance#genetics#decoding dna#genetic code#science#double helix
128 notes
·
View notes
Text
Since day one of COVID in March 2020, I’ve had an issue with referring to the COVID-19 jabs as "gene therapy,” which suggests they provide a benefit. Webster’s defines THERAPY as:
A medical treatment intended to relieve or heal a disorder, injury, or disease.
Consider the following:
Physical therapy, speech therapy, respiratory therapy, art therapy, family therapy, and occupational therapy. These are all ways to improve a health concern.
To the contrary, it has become more obvious every day that these shots were not designed to relieve or heal anything. They certainly didn’t make people healthier. In fact, they have only caused harm and led to increased disease and death.
Gene therapy is a medical treatment designed to intentionally alter a person's genes to treat or cure disease. It involves delivering modified or corrected DNA (or sometimes RNA) into a patient's cells to change how the cells work.
11 notes
·
View notes
Note
i love that u love rabies puppybong...
it’s so fascinating and the craziest part about it is that the entire thing is just 5 different proteins and 1 strand of RNA and the virus is shaped like a bullet. Like holy moly
And ok let’s say you get bit on the leg by a rabid dog. Ok? What happens is the viral particles in the infected saliva will bind to your acetylcholine receptors and worm its way into your muscle cells and start to replicate. And your immune system can’t do shit cuz it’s not like it’s going to attack your own cells. Because it doesn’t even know what’s going on. and it travels from your muscle cells up your nerves and eventually makes its way to your central nervous system and to your brain. It basically hitches a ride inside your neurons. The incubation period is 1-3 months (unless you get bit somewhere with a lot of nerves like your face, then it may be shorter) so you literally have no idea this is happening until it’s too late. Like you are fucked. The 5 stages of the rabies virus are incubation, prodrome (the buildup to the disease stage), acute neurological symptoms, coma, and then death. Once you start showing symptoms you have about a week left to live like it’s over for you.
The 2 forms of rabies are furious (self explanatory, 70-80% of cases) and paralytic (20-30%), where it essentially paralyzes you. Furious form you die within 5-7 days of showing symptoms, paralytic is about 11.
I think most people know the symptoms of rabies (fear of light, fear of water, foamy mouth, etc) but the biology behind it is incredible. Basically what happens is once the virus is in your brain, it travels to your salivary glands and the muscles and the nerves there, and when you try to swallow, you get extremely painful muscle spasming. This is because it replicates in your saliva and swallowing it would just wash it all away (which is where the foaming mouth comes from, you literally cannot swallow). That’s also where the hydrophobia comes from. I couldn’t tell you how it evolved to this behavior but it’s so cool.
There’s also extreme behavior modification like aggression because it fucks with your molecules in such a way you stop producing serotonin. Your brain is just so full of virus you dont know where you end and rabies begins. It takes away your body’s ability to send T cells to help kill the bacteria so it just keeps multiplying and multiplying. then you get viral encephalitis because your brain is literally swollen with rabies and you have dead cells that just keep dying and piling up. And then you die. It’s so cracked
sources: 1 2 3
#ask#sorry i got really excited i hope this is coherent#theres a lot more im neglecting to talk about cuz this post would be miles long but its soooooooooo crazy
7 notes
·
View notes
Text
Genetic engineering: CRISPR and beyond
In genetic engineering, we find ourselves amidst a scientific revolution with the advent of revolutionary technologies like CRISPR-Cas9. However, our journey into the intricate landscape of genetic manipulation is far from complete. This post delves into the nuanced world of genetic engineering, exploring cutting-edge technologies and their remarkable potential in shaping the future of medicine and biotechnology.
CRISPR-Cas9: Precision at the Molecular Level
CRISPR-Cas9, a revolutionary genome editing tool, stands for Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein 9. It utilizes a guide RNA (gRNA) to target specific DNA sequences, and the Cas9 protein acts as molecular scissors to cut the DNA at precisely defined locations. This break in the DNA prompts the cell's natural repair machinery to make changes, either through non-homologous end joining (NHEJ) or homology-directed repair (HDR). CRISPR-Cas9's precision allows for gene knockout, modification, or insertion with remarkable accuracy.
Beyond CRISPR: Emerging Technologies
While CRISPR-Cas9 has dominated the field of genetic engineering, numerous promising technologies have emerged on the horizon. These include CRISPR-Cas variants like CRISPR-Cas12 and CRISPR-Cas13, which offer unique advantages such as smaller size, increased specificity, and targeting of RNA. Additionally, base editing techniques, such as adenine base editors (ABEs) and cytosine base editors (CBEs), enable the direct conversion of one DNA base into another without causing double-strand breaks, expanding the range of genetic modifications possible.
Applications in Medicine
The implications of these advancements are profound, particularly in medicine. Genetic engineering can potentially treat various genetic disorders, from cystic fibrosis to sickle cell anemia, by correcting disease-causing mutations at their source. Precision medicine, tailored to an individual's genetic makeup, is becoming increasingly feasible, allowing for personalized therapies with minimal side effects.
Ethical Considerations and Regulation
As we venture further into the genetic frontier, we must acknowledge the ethical considerations surrounding genetic engineering. The ability to modify the human germline, with implications for future generations, raises ethical dilemmas that necessitate rigorous oversight and regulation. The international community is developing guidelines to ensure responsible use of these powerful tools.
Future Directions and Challenges
While genetic engineering offers immense promise, it is not without its challenges. Off-target effects, unintended consequences, and the potential for creating designer babies are among the issues that demand careful consideration. Researchers and ethicists must work in tandem to navigate this uncharted territory.
References
Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., … & Liu, D. R. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 576(7785), 149-157.
Kime, E. (2021). CRISPR and the ethics of gene editing. Nature Reviews Genetics, 22(1), 3-4.
This post only scratches the surface of the profound transformations occurring in genetic engineering. The relentless pursuit of knowledge and ethical exploration will shape the future of this field as we continue to unlock the intricate secrets of our genetic code.

#science#biology#college#education#school#student#medicine#doctors#health#healthcare#genetics#genetic engineering#crispr#ethical genetics
77 notes
·
View notes
Note
Do you think there's a possibility that Sephiroth has 4 balls rather than the usual pair?
No idea who this person is… but I actually do have an answer. And that’s no.
Because, like, true that Sephiroth was the guy with the highest amount of Jenova cells in him. But all that is like, a graft…? It’s secondary to his biology, not primary. Sephiroth is wholly a human with a few quirks here and there, as well as some crazy post-gestation modifications. He’s got a solidly human base, with additional features. It’s true that Lucrecia was injected with J-cells during pregnancy… but that doesn’t change Sephiroths status as a human. (In real world biology, at least.) all that stuff is secondary
I don’t think he’d have four balls because as far as I know, there are no recorded instances of that ever happening. The only case I could see this happening in is with chimerism. There are no intersex conditions (afaik) where someone has more balls than regulated, and the idea of that happening is pretty crazy considering the sheer amount of checkpoints your DNA and RNA have to hit before your gonads are determined. Anyways gn
15 notes
·
View notes
Text

1917 04 Red Nose Pup - Russell Smith
According to legend, the well-respected Sopwith Pup evolved from a design which test pilot Harry Hawker drew on the Sopwith factory floor. After several modifications this design came to fruition as prototype 3691, making an immediate impression on both the RNAS and the RCF with its speed and maneuverability. The type was pressed into production and given the names "Sopwith "Scout" (RFC) and "Admirality 9901" (RNAS). However, the type became known as the "Pup" after an RFC offer spotted the the Scout sitting next to a 1 1/2 Strutter and declared to the pilot "Good God! Your Strutter has had a pup!" Despite only being armed with a single Vickers machine gun, its light weight and maneuverability made the Pup a good match for the more heavily armed German fighters of the day.This particular Sopwith Pup, N6205, was flown by Flt Cdr Joe Fall of 3 Sq RNAS., the joint leading Pup ace. This Pup featured a red cowling and wheel covers and, not visible in this view, the name BETTY painted under the cockpit.
26 notes
·
View notes
Note
Hello! I am a molecular biologist, and I was wondering if I could get your opinion on some of my theories on Gallifreyans.
I haven't read through everything on your blog yet, but I'm working my way through it (lol). So some of this may not be quite accurate with what you have set up thus far.
Basically, I want to briefly discuss alternative splicing! Anyway, in metazoans, alternative splicing outcomes can be regulated in a time and tissue specific manner by legitimately hundreds of biomolecules such as RNA binding proteins, chromatin remodelers, hormones, etc etc. It is subject to epigenetic regulation as alternative splicing and transcription are coupled (and splicing largely occurs cotranscriptionally), so details such as DNA methylation, nucleosome positioning, histone modifications, etc can change the balance of different mRNA isoforms. This is largely because these factors will either help recruit splicing factors (or inhibit their recruitment) or because it will slow RNA Polymerase II elongation.
Onto my theories. I have been thinking for a little while that the lindos hormone can perhaps modulate splicing, triggering the production of regeneration-specific isoforms. Perhaps their bodies work so fast that isoforms promoting totipotency trigger a temporary transition away from the cells' differentiated states.
I also think it could be possible that they have some novel ability to, say, "unsplice," which humans cannot do. This could potentially allow them to use already made transcripts and then completely change them to produce unique proteins without needing to transcribe another mRNA. This could feasibly allow them to rapidly change what proteins are in each cell (perhaps quick enough that it occurs within the regeneration itself). Although, there would be some instability while now unused proteins get degraded or the splicing/unsplicing ratio stabilizes (the molding period). This would require intense regulation as well as unsplicing and resplicing would now be posttranscriptional, but I digress.
Sorry to bother you with the long post, I just had too many nerdy ideas going through my head. Thanks!
-gallifreyanhotfive
Molecular Biology: 'Unsplicing'
Oh, you thrill me with your biology talk! Molecular biology is not a speciality so apologies in advance for any limited response.
🔬 Lindos and Its Variations
Something to be covered in the new Anatomy and Physiology guide is a wider look at the role of Lindos in Time Lords, so we're hitting the nail on the head here.
Under stress, injury, or during the process of regeneration, the lindal gland significantly increases its production of the hormone Lindoneogen like a caffeine-fueled scientist, resulting in a corresponding surge in lindos cell production. There are several forms of lindos cells, including:
Lindopoetic Progenitor Cells (LPCs): Dormant cells that spring into action upon Lindoneogen stimulation.
Lindopoietic Stem Cells (LSCs): Residing in the yellow bone marrow, ready to differentiate under the guidance of Lindoneogen and the catalytic influence of artron, into ...
Lindoblasts and Phagolindotropes: Specialised cells responsible for regenerating tissue and recycling cellular components from the previous incarnation.
Haemolindocytes: Circulating cells that endow Gallifreyan blood with its regenerative properties.
💡Splicing and Lindoneogen
Lindoneogen could play a key role in alternative splicing, creating specific mRNA isoforms vital for regeneration. This implies that Lindoneogen is not just a cellular signal but also a molecular tool for crafting the necessary protein portfolio for regeneration. So Lindoneogen may trigger the production of specific mRNA isoforms that are vital for the regeneration process, which could lead to the expression of proteins that facilitate the transition of cells into a more pluripotent state.
🖇️Unsplicing
Love this idea. 'Unsplicing' as your concept presents would be particularly relevant during regeneration. It could allow cells to quickly alter their protein expression profiles without the lag of new mRNA transcription. This rapid adaptation would be pretty handy for the efficient transition of cells to suit the requirements of the new incarnation.
🔗Integrating with Lindos Cells
This concept of 'unsplicing' could be particularly prominent in the function of phagolindotropes. As these cells are responsible for consuming the previous incarnation's cells and replacing them with new ones, their ability to 'unsplice' and rapidly change protein expression would be pretty useful. This mechanism might also support the functions of lindoblasts and haemolindocytes in tissue regeneration and blood adaptability.
🏫 So ...
The addition of splicing and unsplicing mechanisms to the lindos theory suggests a more complex and dynamic process than simple cellular proliferation and differentiation, with dynamic genetic adaptations at the molecular level highlighting the advanced biological capabilities of Gallifreyans. Good work, Batman!
Any orange text is educated guesswork or theoretical. More content ... →📫Got a question? | 📚Complete list of Q+A and factoids →📢Announcements |🩻Biology |🗨️Language |🕰️Throwbacks |🤓Facts → Features:⭐Guest Posts | 🍜Chomp Chomp with Myishu →🫀Gallifreyan Anatomy and Physiology Guide (pending) →⚕️Gallifreyan Emergency Medicine Guides →📝Source list (WIP) →📜Masterpost If you're finding your happy place in this part of the internet, feel free to buy a coffee to help keep our exhausted human conscious. She works full-time in medicine and is so very tired 😴
#doctor who#gil#gallifrey institute for learning#dr who#dw eu#gallifrey#gil biology#gallifreyans#ask answered#GIL: Asks#gallifreyan biology#GIL: Biology#GIL: Biology/Foundations#GIL: Species/Gallifreyans
21 notes
·
View notes
Text
DNA Methylation
Hello! This week's educational post is on DNA methylation. Note that DNA methylation functions a bit differently in plants vs animals. This post will be more plant-centric since that is what I study.
DNA methylation refers to the addition of a methyl group (one carbon molecule) to the 5' position of a cytosine base in the DNA. This is a type of epigenetic mark, or a heritable genetic modification without a change in DNA sequence (Takuno & Gaut, 2012). It alters the expression of genes through transcriptional silencing, usually by preventing the binding of polymerase. However, the exact ways methylation regulates and is regulated can differ depending on context (Neiderhuth & Schmitz, 2016). There are three contexts in which DNA methylation can occur: CG, CHG, and CHH, where H represents A, T, or C (Neiderhuth & Schmitz, 2016).
Methylation in the CG context, known as CpG methylation, is the most stable of the three contexts. After DNA replication occurs, VIM1-5 recognizes hemi-methylated CG sites and recruits the methyltransferase MET1 to methylate the unmethylated strand (Neiderhuth & Schmitz, 2016). CpG methylation around the transcriptional start site causes transcriptional silencing, while CpG methylation in the gene body is associated with active, and even increased, transcription (Neiderhuth & Schmitz, 2016).
Methylation in the CHG context is the second most stable. It is maintained by the methyltransferase CMT3, which depends on the H3K9me2 histone modification (I'll go over histone modifications for my next educational post) in a positive feedback loop that maintains the locations of both these epigenetic marks (Neiderhuth & Schmitz, 2016). This feedback loop can be broken by the histone demethylase IBM1, causing transcription to become reactivated (Neiderhuth & Schmitz, 2016).
CHH methylation cannot be maintained after DNA replication due to its asymmetry: lack of a guanine on the methylated strand means there is no cytosine to methylate on the opposite strand (Neiderhuth & Schmitz, 2016). CHH methylation is always established de novo by the methyltransferase CMT2 or through the RNA-directed DNA methylation (RdDM) pathway (Neiderhuth & Schmitz, 2016).
De novo DNA methylation for all three contexts can be mediated by the RdDM pathway (Zhang et al, 2018). In this process, short-interfering RNAs (siRNAs) work together with scaffold RNAs and a variety of proteins to guide the methyltransferase DRM2 to target regions for methylation (Zhang et al, 2018).
Transcriptional regulation through DNA methylation is important for a variety of processes related to plant development and genome stability. Specifically, it has been shown to be involved in transposon silencing (Slotkin & Martienssen, 2007), fruit ripening (Xiao et al, 2020), genetic imprinting (Rodrigues et al, 2013), and aging (D’Amico-Willman et al, 2024).
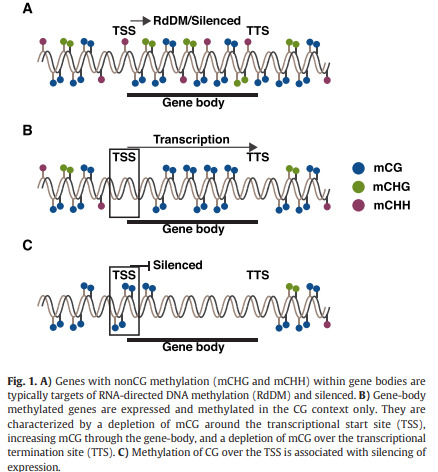
(Neiderhuth & Schmitz, 2016)
Important Terms: DNA methylation, epigenetics, cytosine, transcription, methyltransferase
6 notes
·
View notes
Text
Navigating the Complexity of Alternative Splicing in Eukaryotic Gene Expression: A Molecular Odyssey
Embarking on the journey of molecular biology exposes students to the marvels and intricacies of life at the molecular level. One captivating aspect within this domain is the phenomenon of alternative splicing, where a single gene orchestrates a symphony of diverse protein isoforms. As students grapple with questions related to this molecular intricacy, the role of a reliable molecular biology Assignment Helper becomes indispensable. This blog delves into a challenging question, exploring the mechanisms and consequences of alternative splicing, shedding light on its pivotal role in molecular biology.
Question: Explain the mechanisms and consequences of alternative splicing in eukaryotic gene expression, highlighting its role in generating proteomic diversity and the potential impact on cellular function. Additionally, discuss any recent advancements or discoveries that have provided insight into the regulation and functional significance of alternative splicing.
Answer: Alternative splicing, a maestro in the grand composition of gene expression, intricately weaves the fabric of molecular diversity. Mechanistically, this phenomenon employs exon skipping, intron retention, and alternative 5' or 3' splice sites to sculpt multiple mRNA isoforms from a single gene.
The repercussions of alternative splicing resonate deeply within the proteomic landscape. Proteins, diverse in function, emerge as a consequence, adding layers of complexity to cellular processes. Tissue-specific expression, another outcome, paints a vivid picture of the nuanced orchestration of cellular differentiation.
Regulating this intricate dance of alternative splicing involves an ensemble cast of splicing factors, enhancers, silencers, and epigenetic modifications. In the ever-evolving landscape, recent breakthroughs in high-throughput sequencing techniques, notably RNA-seq, offer a panoramic view of splicing patterns across diverse tissues and conditions. CRISPR/Cas9 technology, a molecular tool of precision, enables the manipulation of splicing factor expression, unraveling their roles in the intricate regulation of alternative splicing.
In the dynamic realm of molecular biology, alternative splicing emerges as a linchpin. Specific splicing events, linked to various diseases, beckon researchers towards therapeutic interventions. The complexities embedded in this molecular tapestry underscore the perpetual need for exploration and comprehension.
Conclusion: The odyssey through alternative splicing unveils its prominence as a cornerstone in the narrative of molecular biology. From sculpting proteomic diversity to influencing cellular functions, alternative splicing encapsulates the essence of molecular intricacies. For students navigating this terrain, the exploration of questions like these not only deepens understanding but also propels us into a realm of limitless possibilities.
#molecular biology assignment help#biology assignment help#university#college#assignment help#pay to do assignment
9 notes
·
View notes
Text
Abstract
Therapeutic applications of synthetic mRNA were proposed more than 30 years ago, and are currently the basis of one of the vaccine platforms used at a massive scale as part of the public health strategy to get COVID-19 under control. To date, there are no published studies on the biodistribution, cellular uptake, endosomal escape, translation rates, functional half-life and inactivation kinetics of synthetic mRNA, rates and duration of vaccine-induced antigen expression in different cell types. Furthermore, despite the assumption that there is no possibility of genomic integration of therapeutic synthetic mRNA, only one recent study has examined interactions between vaccine mRNA and the genome of transfected cells, and reported that an endogenous retrotransposon, LINE-1 is unsilenced following mRNA entry to the cell, leading to reverse transcription of full length vaccine mRNA sequences, and nuclear entry. This finding should be a major safety concern, given the possibility of synthetic mRNA-driven epigenetic and genomic modifications arising. We propose that in susceptible individuals, cytosolic clearance of nucleotide modified synthetic (nms-mRNAs) is impeded. Sustained presence of nms-mRNA in the cytoplasm deregulates and activates endogenous transposable elements (TEs), causing some of the mRNA copies to be reverse transcribed. The cytosolic accumulation of the nms-mRNA and the reverse transcribed cDNA molecules activates RNA and DNA sensory pathways. Their concurrent activation initiates a synchronized innate response against non-self nucleic acids, prompting type-I interferon and pro-inflammatory cytokine production which, if unregulated, leads to autoinflammatory and autoimmune conditions, while activated TEs increase the risk of insertional mutagenesis of the reverse transcribed molecules, which can disrupt coding regions, enhance the risk of mutations in tumour suppressor genes, and lead to sustained DNA damage. Susceptible individuals would then expectedly have an increased risk of DNA damage, chronic autoinflammation, autoimmunity and cancer. In light of the current mass administration of nms-mRNA vaccines, it is essential and urgent to fully understand the intracellular cascades initiated by cellular uptake of synthetic mRNA and the consequences of these molecular events.
4 notes
·
View notes